Os06g0685100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to COBRA-like protein 10 precursor.

FiT-DB / Search/ Help/ Sample detail
|
Os06g0685100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to COBRA-like protein 10 precursor.
|
|
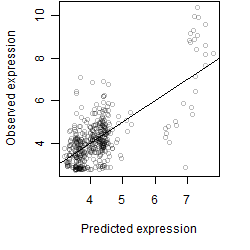
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable
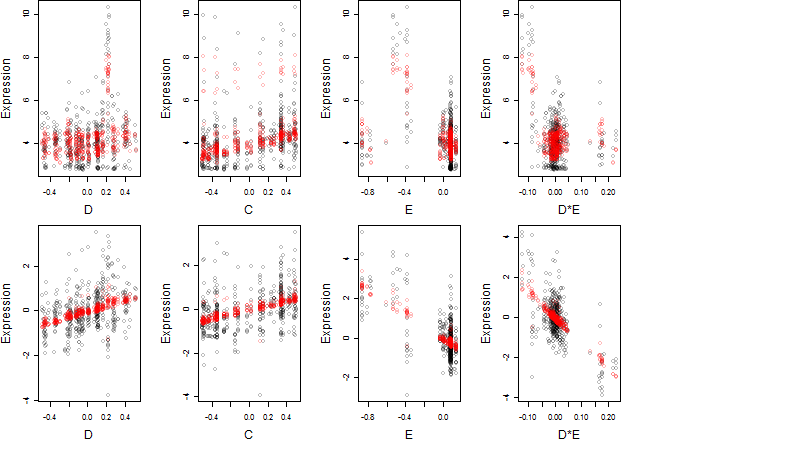
Residual plot

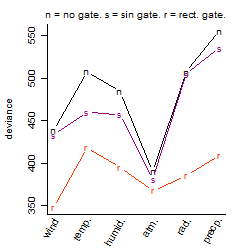
Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = < th, dose dependency = dose independent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 345.28 | 0.873 | 4.31 | 1.16 | 0.963 | -2.84 | 1.14 | -12.1 | -0.263 | 15 | 6.413 | 4322 | 19.2 | 3.14 |
| 462.76 | 1 | 4.31 | 1.05 | 1.02 | -1.92 | 0.597 | -- | -0.242 | 14.9 | 8.099 | 4322 | 19.2 | 3.09 |
| 344.79 | 0.865 | 4.31 | 1.22 | 1.02 | -3.02 | -- | -12.3 | -0.235 | 15.2 | 6.491 | 4325 | 19.7 | 3.1 |
| 464.79 | 1 | 4.31 | 1 | 1 | -1.89 | -- | -- | -0.22 | 14.9 | 8.081 | 4322 | 19.2 | 3.09 |
| 402.78 | 0.935 | 4.31 | 1.22 | -- | -2.95 | -- | -12.3 | -0.235 | -- | 6.491 | 4325 | 19.7 | 3.1 |
| 583.01 | 1.13 | 4.27 | 0.913 | 1 | -- | 1.09 | -- | -0.415 | 14.7 | -- | -- | -- | -- |
| 587.2 | 1.13 | 4.27 | 0.94 | 1 | -- | -- | -- | -0.408 | 14.7 | -- | -- | -- | -- |
| 489.31 | 1.03 | 4.29 | 1.97 | -- | -2.08 | -- | -- | -0.0579 | -- | 5.7 | 10889 | 19.3 | 2.42 |
| 486.43 | 1.03 | 4.31 | -- | 0.984 | -1.98 | -- | -- | -0.22 | 15 | 8.092 | 4322 | 19.2 | 3.12 |
| 646.09 | 1.19 | 4.28 | 0.897 | -- | -- | -- | -- | -0.434 | -- | -- | -- | -- | -- |
| 610.5 | 1.16 | 4.29 | -- | 0.986 | -- | -- | -- | -0.513 | 14.6 | -- | -- | -- | -- |
| 522.59 | 1.06 | 4.28 | -- | -- | -2.49 | -- | -- | -0.0522 | -- | 5.6 | 10901 | 19.3 | 2.42 |
| 667.34 | 1.21 | 4.3 | -- | -- | -- | -- | -- | -0.533 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance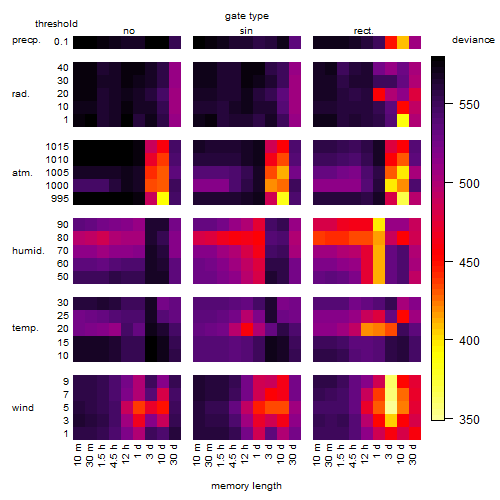
|
Histogram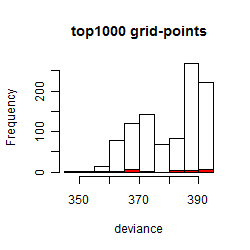 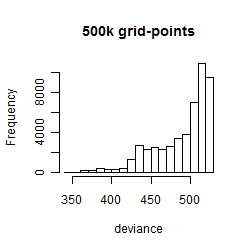
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 348.45 | wind | 5 | 4320 | > th | dose dependent | rect. | 20 | 3 |
| 5 | 356.86 | wind | 7 | 4320 | < th | dose independent | rect. | 19 | 3 |
| 16 | 359.59 | wind | 7 | 4320 | > th | dose independent | rect. | 20 | 2 |
| 32 | 360.63 | wind | 9 | 4320 | > th | dose dependent | rect. | 19 | 7 |
| 119 | 368.13 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 15 | 3 |
| 120 | 368.13 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 10 | 10 |
| 122 | 368.16 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 7 | 13 |
| 124 | 368.18 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 15 | 5 |
| 141 | 368.50 | atmosphere | 995 | 14400 | < th | dose independent | rect. | 15 | 3 |
| 205 | 369.15 | atmosphere | 995 | 14400 | < th | dose dependent | rect. | 15 | 4 |
| 329 | 372.60 | atmosphere | 995 | 14400 | < th | dose independent | rect. | 18 | 1 |
| 332 | 372.60 | atmosphere | 995 | 14400 | < th | dose dependent | rect. | 18 | 1 |
| 442 | 381.35 | atmosphere | 995 | 14400 | > th | dose independent | sin | 22 | NA |
| 453 | 381.76 | atmosphere | 995 | 14400 | < th | dose independent | sin | 22 | NA |
| 481 | 383.83 | atmosphere | 995 | 14400 | < th | dose dependent | sin | 22 | NA |
| 485 | 384.05 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 20 | 20 |
| 513 | 385.14 | radiation | 1 | 14400 | < th | dose dependent | rect. | 20 | 6 |
| 596 | 387.05 | radiation | 1 | 14400 | < th | dose dependent | rect. | 23 | 1 |
| 609 | 387.49 | radiation | 1 | 14400 | < th | dose dependent | rect. | 22 | 4 |
| 801 | 390.20 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 14 | 23 |
| 802 | 390.21 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 12 | 23 |
| 804 | 390.22 | atmosphere | 995 | 14400 | > th | dose independent | no | NA | NA |
| 805 | 390.23 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 10 | 23 |
| 874 | 391.33 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 14 | 21 |
| 920 | 391.96 | atmosphere | 995 | 14400 | < th | dose independent | no | NA | NA |