Os06g0486800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Formate dehydrogenase, mitochondrial precursor (EC 1.2.1.2) (NAD- dependent formate dehydrogenase) (FDH).
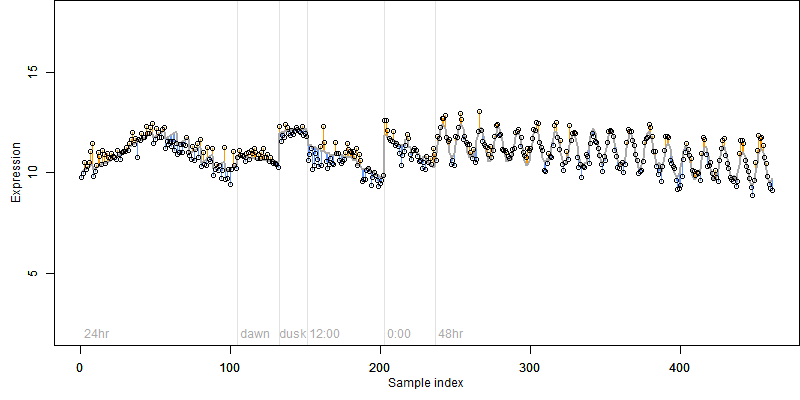

FiT-DB / Search/ Help/ Sample detail
|
Os06g0486800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Formate dehydrogenase, mitochondrial precursor (EC 1.2.1.2) (NAD- dependent formate dehydrogenase) (FDH).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable

Residual plot

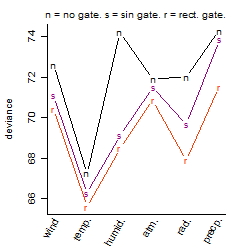
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 66.82 | 0.384 | 11.1 | -1.62 | 1.7 | 0.716 | 0.6 | -0.0308 | -0.176 | 19.2 | 25.6 | 1340 | -- | -- |
| 66.8 | 0.381 | 11.1 | -1.61 | 1.7 | 0.708 | 0.596 | -- | -0.165 | 19.2 | 25.8 | 1329 | -- | -- |
| 67.75 | 0.383 | 11.1 | -1.61 | 1.69 | 0.707 | -- | 0.0376 | -0.164 | 19.1 | 25.77 | 1306 | -- | -- |
| 67.74 | 0.383 | 11.1 | -1.62 | 1.69 | 0.716 | -- | -- | -0.164 | 19.1 | 25.79 | 1300 | -- | -- |
| 133.94 | 0.539 | 11.1 | -1.92 | -- | 2 | -- | -0.921 | -0.383 | -- | 24 | 548 | -- | -- |
| 87.96 | 0.44 | 11.1 | -1.37 | 1.68 | -- | 0.55 | -- | -0.111 | 19.4 | -- | -- | -- | -- |
| 88.79 | 0.441 | 11.1 | -1.39 | 1.68 | -- | -- | -- | -0.11 | 19.4 | -- | -- | -- | -- |
| 134.72 | 0.541 | 11.1 | -1.78 | -- | 1.91 | -- | -- | -0.345 | -- | 23.98 | 552 | -- | -- |
| 131.28 | 0.534 | 11 | -- | 1.69 | 0.39 | -- | -- | 0.098 | 19.2 | 28.22 | 1234 | -- | -- |
| 238.63 | 0.722 | 11 | -1.41 | -- | -- | -- | -- | -0.137 | -- | -- | -- | -- | -- |
| 139.39 | 0.552 | 11 | -- | 1.69 | -- | -- | -- | 0.0443 | 19.3 | -- | -- | -- | -- |
| 214.86 | 0.683 | 11 | -- | -- | 1.61 | -- | -- | -0.0919 | -- | 24.5 | 524 | -- | -- |
| 291.29 | 0.797 | 11 | -- | -- | -- | -- | -- | 0.0194 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance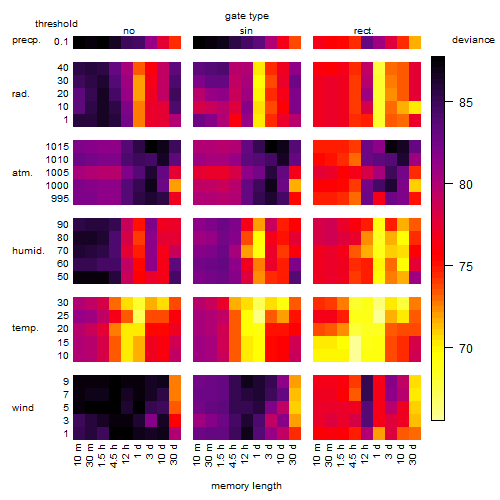
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 65.57 | temperature | 10 | 270 | > th | dose dependent | rect. | 24 | 17 |
| 3 | 65.86 | temperature | 25 | 1440 | > th | dose dependent | rect. | 15 | 6 |
| 9 | 65.92 | temperature | 25 | 1440 | > th | dose dependent | rect. | 15 | 9 |
| 26 | 66.11 | temperature | 25 | 1440 | > th | dose dependent | rect. | 17 | 2 |
| 52 | 66.25 | temperature | 25 | 1440 | > th | dose dependent | sin | 16 | NA |
| 103 | 66.58 | temperature | 30 | 1440 | < th | dose independent | rect. | 14 | 11 |
| 119 | 66.62 | temperature | 30 | 1440 | < th | dose independent | rect. | 14 | 5 |
| 138 | 66.73 | temperature | 30 | 1440 | > th | dose independent | rect. | 14 | 5 |
| 206 | 66.98 | temperature | 30 | 1440 | < th | dose independent | sin | 14 | NA |
| 323 | 67.21 | temperature | 30 | 1440 | > th | dose independent | sin | 14 | NA |
| 335 | 67.22 | temperature | 25 | 1440 | > th | dose dependent | no | NA | NA |
| 354 | 67.23 | temperature | 25 | 1440 | > th | dose dependent | rect. | 23 | 23 |
| 393 | 67.36 | temperature | 25 | 1440 | > th | dose dependent | rect. | 17 | 23 |
| 500 | 67.74 | temperature | 30 | 14400 | > th | dose dependent | rect. | 17 | 2 |
| 554 | 67.88 | radiation | 40 | 1440 | < th | dose dependent | rect. | 17 | 9 |
| 599 | 67.96 | temperature | 30 | 1440 | < th | dose independent | rect. | 17 | 23 |
| 641 | 68.05 | temperature | 30 | 1440 | < th | dose independent | no | NA | NA |
| 643 | 68.05 | temperature | 30 | 1440 | < th | dose independent | rect. | 6 | 14 |
| 662 | 68.05 | temperature | 30 | 1440 | < th | dose independent | rect. | 21 | 22 |
| 685 | 68.06 | temperature | 30 | 1440 | < th | dose independent | rect. | 23 | 20 |
| 728 | 68.12 | temperature | 30 | 270 | < th | dose dependent | rect. | 5 | 18 |
| 773 | 68.24 | radiation | 1 | 1440 | > th | dose dependent | rect. | 16 | 15 |
| 791 | 68.26 | radiation | 1 | 1440 | > th | dose dependent | rect. | 16 | 3 |
| 815 | 68.30 | radiation | 10 | 1440 | > th | dose independent | rect. | 16 | 14 |
| 819 | 68.32 | radiation | 10 | 1440 | > th | dose independent | rect. | 16 | 2 |
| 832 | 68.32 | temperature | 30 | 1440 | > th | dose independent | rect. | 17 | 23 |
| 874 | 68.37 | temperature | 30 | 1440 | > th | dose independent | no | NA | NA |
| 999 | 68.44 | temperature | 30 | 14400 | > th | dose independent | rect. | 17 | 2 |