Os06g0338700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Copper amine oxidase family protein.
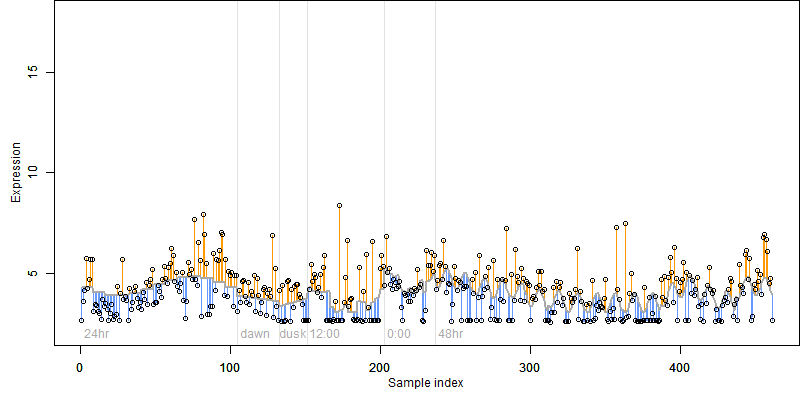

FiT-DB / Search/ Help/ Sample detail
|
Os06g0338700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Copper amine oxidase family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
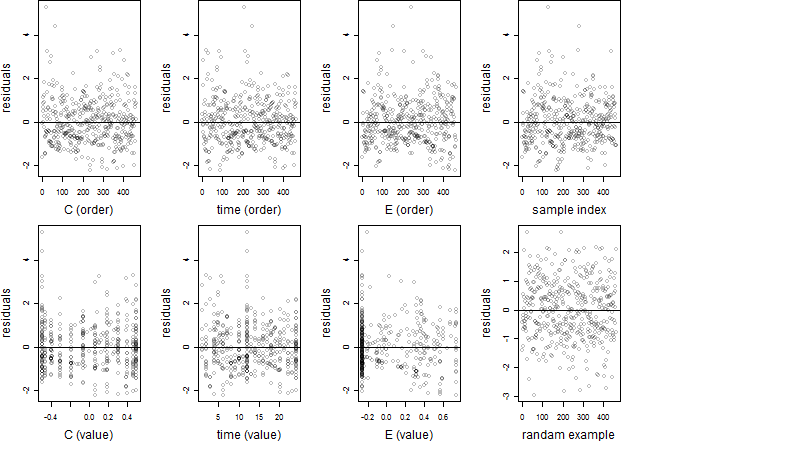
Dependence on each variable

Residual plot
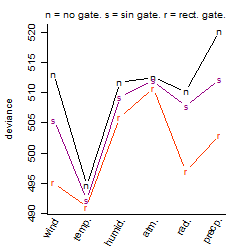
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 493.01 | 1.04 | 3.97 | 0.596 | 0.864 | 2.66 | 0.782 | 3.54 | 0.725 | 0.599 | 19.3 | 4399 | -- | -- |
| 497.49 | 1.04 | 3.84 | 0.854 | 1.01 | 1.75 | 0.945 | -- | 0.946 | 0.523 | 21.7 | 3018 | -- | -- |
| 495.6 | 1.04 | 3.98 | 0.577 | 0.861 | 2.67 | -- | 3.55 | 0.727 | 0.531 | 19.31 | 4411 | -- | -- |
| 496.16 | 1.04 | 3.84 | 0.658 | 0.877 | 1.49 | -- | -- | 0.867 | 0.482 | 21.7 | 3029 | -- | -- |
| 541.7 | 1.08 | 3.97 | 0.607 | -- | 2.71 | -- | 3.66 | 0.731 | -- | 19.21 | 4500 | -- | -- |
| 557.09 | 1.11 | 3.94 | -0.109 | 0.844 | -- | 0.742 | -- | 0.374 | 0.457 | -- | -- | -- | -- |
| 559.43 | 1.11 | 3.95 | -0.137 | 0.841 | -- | -- | -- | 0.371 | 0.372 | -- | -- | -- | -- |
| 545.18 | 1.09 | 3.83 | 0.663 | -- | 1.43 | -- | -- | 0.867 | -- | 21.7 | 3070 | -- | -- |
| 503.02 | 1.04 | 3.83 | -- | 0.955 | 1.22 | -- | -- | 0.828 | 0.501 | 22.4 | 2782 | -- | -- |
| 603.91 | 1.15 | 3.93 | -0.109 | -- | -- | -- | -- | 0.385 | -- | -- | -- | -- | -- |
| 559.93 | 1.11 | 3.94 | -- | 0.839 | -- | -- | -- | 0.387 | 0.363 | -- | -- | -- | -- |
| 553 | 1.1 | 3.84 | -- | -- | 1.13 | -- | -- | 0.777 | -- | 22.4 | 3063 | -- | -- |
| 604.22 | 1.15 | 3.93 | -- | -- | -- | -- | -- | 0.397 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance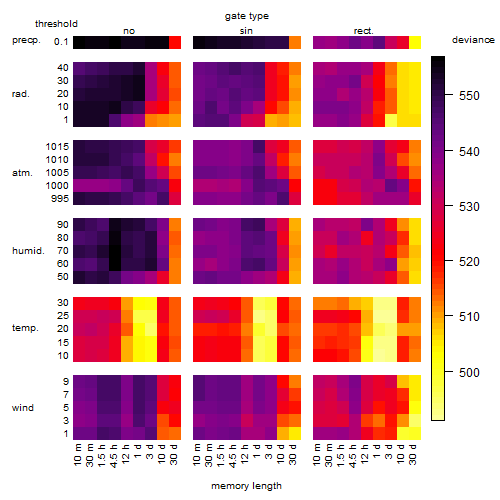
|
Histogram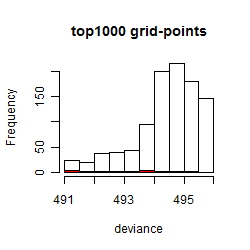 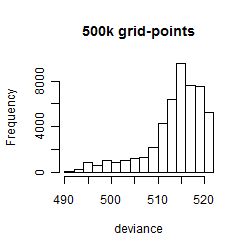
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 491.03 | temperature | 25 | 1440 | < th | dose dependent | rect. | 19 | 12 |
| 3 | 491.09 | temperature | 25 | 1440 | < th | dose dependent | rect. | 19 | 9 |
| 6 | 491.23 | temperature | 25 | 1440 | < th | dose dependent | rect. | 22 | 6 |
| 15 | 491.37 | temperature | 25 | 1440 | < th | dose dependent | rect. | 2 | 1 |
| 34 | 491.84 | temperature | 10 | 1440 | > th | dose dependent | rect. | 2 | 1 |
| 58 | 492.22 | temperature | 25 | 1440 | < th | dose dependent | sin | 12 | NA |
| 72 | 492.39 | temperature | 25 | 1440 | < th | dose dependent | rect. | 12 | 19 |
| 100 | 492.84 | temperature | 25 | 1440 | < th | dose dependent | rect. | 5 | 1 |
| 143 | 493.33 | temperature | 10 | 1440 | > th | dose dependent | rect. | 1 | 6 |
| 152 | 493.41 | temperature | 10 | 1440 | > th | dose dependent | rect. | 23 | 8 |
| 174 | 493.60 | temperature | 10 | 1440 | > th | dose dependent | rect. | 23 | 5 |
| 197 | 493.73 | temperature | 20 | 4320 | < th | dose dependent | rect. | 2 | 22 |
| 207 | 493.77 | temperature | 10 | 1440 | > th | dose dependent | rect. | 19 | 12 |
| 212 | 493.80 | temperature | 15 | 1440 | > th | dose dependent | rect. | 5 | 1 |
| 309 | 494.18 | temperature | 20 | 4320 | < th | dose dependent | rect. | 7 | 22 |
| 394 | 494.40 | temperature | 20 | 4320 | < th | dose dependent | rect. | 19 | 20 |
| 520 | 494.61 | temperature | 20 | 4320 | < th | dose dependent | no | NA | NA |
| 686 | 495.06 | wind | 3 | 43200 | < th | dose dependent | rect. | 22 | 1 |
| 781 | 495.33 | temperature | 20 | 4320 | < th | dose dependent | rect. | 2 | 12 |