Os06g0142400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Early nodulin.


FiT-DB / Search/ Help/ Sample detail
|
Os06g0142400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Early nodulin.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
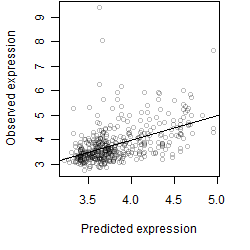 __
__

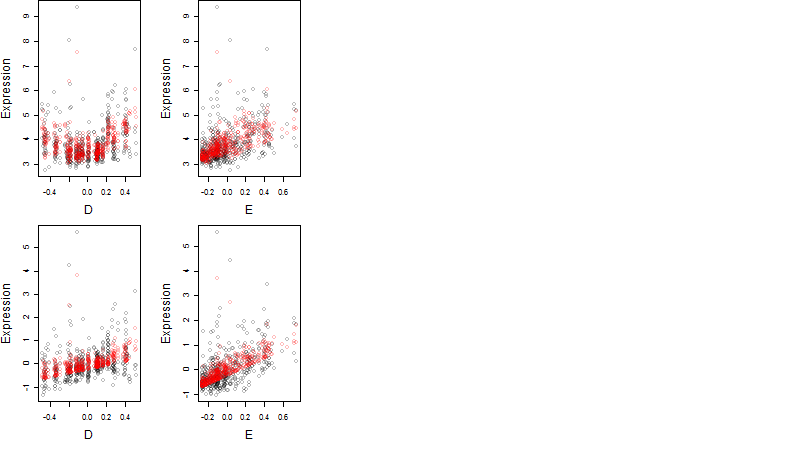
Dependence on each variable
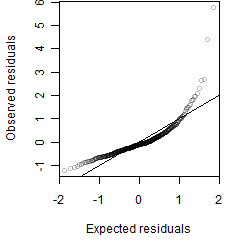
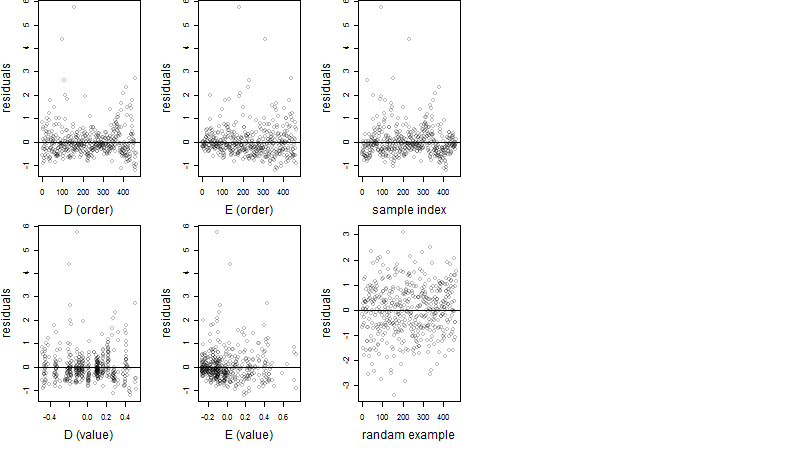
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 188.08 | 0.644 | 3.81 | 0.608 | 0.319 | 1.97 | -0.783 | 1.66 | 0.0411 | 5.92 | 24.1 | 884 | -- | -- |
| 191.21 | 0.644 | 3.77 | 0.87 | 0.308 | 1.75 | -0.74 | -- | 0.114 | 5.47 | 24.88 | 867 | -- | -- |
| 189.52 | 0.641 | 3.81 | 0.604 | 0.323 | 1.96 | -- | 1.64 | 0.0431 | 6.17 | 24.1 | 901 | -- | -- |
| 192.47 | 0.646 | 3.77 | 0.856 | 0.308 | 1.73 | -- | -- | 0.104 | 6.24 | 24.72 | 870 | -- | -- |
| 194.1 | 0.649 | 3.81 | 0.626 | -- | 2 | -- | 1.49 | 0.0593 | -- | 24.15 | 673 | -- | -- |
| 235.76 | 0.72 | 3.85 | 0.37 | 0.43 | -- | -0.174 | -- | -0.248 | 8.1 | -- | -- | -- | -- |
| 235.83 | 0.719 | 3.85 | 0.366 | 0.429 | -- | -- | -- | -0.247 | 8.25 | -- | -- | -- | -- |
| 194.56 | 0.65 | 3.77 | 0.854 | -- | 1.74 | -- | -- | 0.131 | -- | 27.83 | 658 | -- | -- |
| 209.34 | 0.674 | 3.81 | -- | 0.262 | 1.34 | -- | -- | -0.0555 | 7.12 | 24.76 | 672 | -- | -- |
| 246 | 0.733 | 3.86 | 0.369 | -- | -- | -- | -- | -0.242 | -- | -- | -- | -- | -- |
| 239.36 | 0.724 | 3.86 | -- | 0.431 | -- | -- | -- | -0.288 | 8.18 | -- | -- | -- | -- |
| 211.43 | 0.677 | 3.81 | -- | -- | 1.41 | -- | -- | -0.0307 | -- | 28.62 | 608 | -- | -- |
| 249.59 | 0.737 | 3.87 | -- | -- | -- | -- | -- | -0.283 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 187.56 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 14 |
| 2 | 187.78 | temperature | 25 | 720 | < th | dose dependent | rect. | 24 | 21 |
| 10 | 188.66 | temperature | 30 | 720 | < th | dose dependent | rect. | 17 | 19 |
| 13 | 188.74 | temperature | 25 | 1440 | < th | dose dependent | rect. | 16 | 6 |
| 53 | 189.49 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 20 |
| 75 | 189.75 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 89 | 189.98 | temperature | 25 | 720 | < th | dose dependent | rect. | 8 | 23 |
| 140 | 190.58 | temperature | 25 | 1440 | < th | dose dependent | sin | 17 | NA |
| 141 | 190.58 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 2 |
| 146 | 190.62 | temperature | 25 | 720 | < th | dose dependent | rect. | 3 | 23 |
| 151 | 190.64 | temperature | 10 | 720 | > th | dose dependent | rect. | 12 | 21 |
| 204 | 191.21 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 6 |
| 249 | 191.49 | temperature | 25 | 720 | < th | dose dependent | sin | 10 | NA |
| 250 | 191.49 | temperature | 10 | 1440 | > th | dose dependent | rect. | 21 | 1 |
| 367 | 192.10 | temperature | 10 | 720 | > th | dose dependent | sin | 10 | NA |
| 602 | 193.00 | temperature | 10 | 1440 | > th | dose dependent | sin | 14 | NA |
| 623 | 193.07 | wind | 9 | 43200 | > th | dose dependent | rect. | 18 | 12 |
| 742 | 193.39 | temperature | 20 | 720 | > th | dose independent | rect. | 3 | 20 |