Os05g0400700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to DNA-directed RNA polymerase II 19 kDa polypeptide (EC 2.7.7.6) (RNA polymerase II subunit 5).
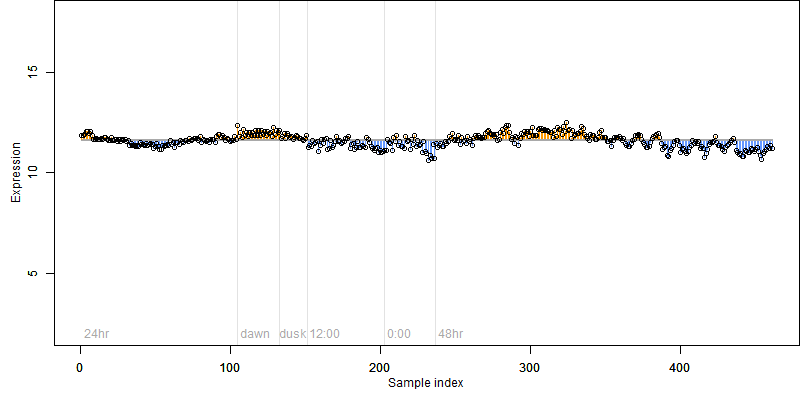

FiT-DB / Search/ Help/ Sample detail
|
Os05g0400700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to DNA-directed RNA polymerase II 19 kDa polypeptide (EC 2.7.7.6) (RNA polymerase II subunit 5).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
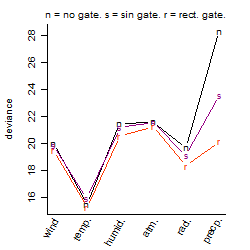
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15.24 | 0.183 | 11.7 | -1.16 | 0.699 | 1.18 | 0.066 | -1.65 | -0.343 | 6.39 | 16.15 | 557 | -- | -- |
| 16.86 | 0.191 | 11.6 | -0.864 | 0.678 | 1.2 | 0.455 | -- | -0.258 | 6.49 | 14.98 | 574 | -- | -- |
| 15.24 | 0.182 | 11.7 | -1.17 | 0.703 | 1.18 | -- | -1.7 | -0.346 | 6.35 | 16.2 | 550 | -- | -- |
| 17.33 | 0.194 | 11.6 | -0.866 | 0.689 | 1.21 | -- | -- | -0.261 | 6.44 | 13.96 | 577 | -- | -- |
| 24.14 | 0.229 | 11.7 | -1.21 | -- | 0.929 | -- | -1.04 | -0.323 | -- | 12.75 | 2708 | -- | -- |
| 34.65 | 0.276 | 11.6 | -0.58 | 0.387 | -- | 0.178 | -- | -0.0766 | 5.81 | -- | -- | -- | -- |
| 34.72 | 0.276 | 11.6 | -0.579 | 0.388 | -- | -- | -- | -0.0774 | 5.74 | -- | -- | -- | -- |
| 23.8 | 0.227 | 11.7 | -0.894 | -- | 1.05 | -- | -- | -0.259 | -- | 12.15 | 1319 | -- | -- |
| 34.74 | 0.275 | 11.6 | -- | 0.603 | 0.835 | -- | -- | -0.116 | 6.04 | 13.76 | 466 | -- | -- |
| 42.51 | 0.305 | 11.6 | -0.567 | -- | -- | -- | -- | -0.0682 | -- | -- | -- | -- | -- |
| 43.56 | 0.309 | 11.6 | -- | 0.378 | -- | -- | -- | -0.0129 | 5.81 | -- | -- | -- | -- |
| 42.09 | 0.302 | 11.6 | -- | -- | 0.675 | -- | -- | -0.104 | -- | 8.606 | 1283 | -- | -- |
| 50.99 | 0.333 | 11.6 | -- | -- | -- | -- | -- | -0.00533 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance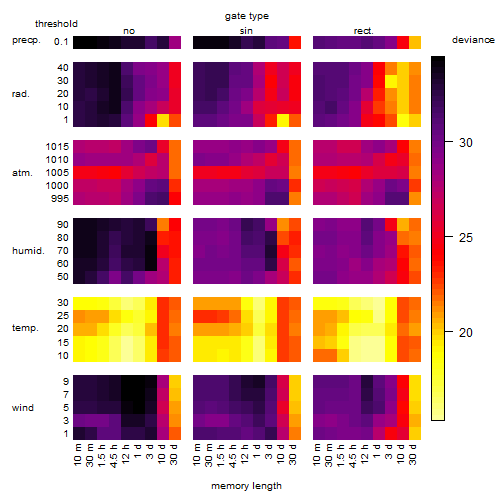
|
Histogram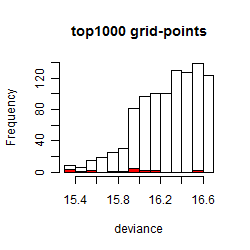 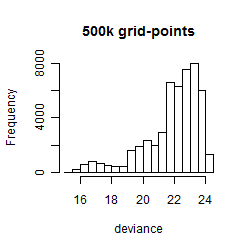
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 15.30 | temperature | 10 | 1440 | > th | dose dependent | rect. | 22 | 1 |
| 2 | 15.32 | temperature | 15 | 720 | > th | dose dependent | rect. | 10 | 22 |
| 8 | 15.38 | temperature | 30 | 1440 | < th | dose dependent | rect. | 22 | 1 |
| 10 | 15.45 | temperature | 10 | 720 | > th | dose dependent | rect. | 1 | 23 |
| 15 | 15.52 | temperature | 15 | 720 | > th | dose dependent | no | NA | NA |
| 27 | 15.58 | temperature | 15 | 720 | > th | dose dependent | rect. | 17 | 23 |
| 66 | 15.78 | temperature | 15 | 720 | > th | dose dependent | rect. | 23 | 23 |
| 91 | 15.88 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 14 |
| 118 | 15.92 | temperature | 30 | 720 | < th | dose dependent | rect. | 14 | 23 |
| 125 | 15.94 | temperature | 30 | 720 | < th | dose dependent | rect. | 14 | 21 |
| 141 | 15.97 | temperature | 10 | 1440 | > th | dose dependent | sin | 12 | NA |
| 164 | 15.99 | temperature | 30 | 1440 | < th | dose dependent | rect. | 17 | 14 |
| 183 | 16.00 | temperature | 30 | 720 | < th | dose dependent | rect. | 6 | 23 |
| 214 | 16.04 | temperature | 30 | 720 | < th | dose dependent | no | NA | NA |
| 265 | 16.09 | temperature | 30 | 720 | < th | dose dependent | rect. | 22 | 23 |
| 303 | 16.13 | temperature | 30 | 1440 | < th | dose dependent | sin | 12 | NA |
| 325 | 16.14 | temperature | 30 | 720 | < th | dose dependent | rect. | 3 | 23 |
| 772 | 16.53 | temperature | 10 | 1440 | > th | dose dependent | rect. | 4 | 3 |
| 854 | 16.59 | temperature | 30 | 1440 | < th | dose dependent | rect. | 4 | 3 |