Os05g0245300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF588 family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os05g0245300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF588 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
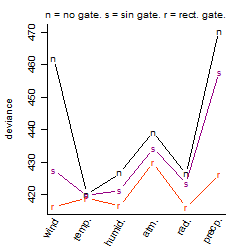
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = radiation,
response mode = < th, dose dependency = dose dependent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 421.22 | 0.964 | 3.98 | -3.17 | 0.256 | -0.622 | 0.936 | 7.83 | -0.397 | 2.08 | 1.028 | 17019 | 5.01 | 0.951 |
| 492.9 | 1.03 | 3.93 | -1.63 | 0.289 | -1.54 | 0.835 | -- | -0.0897 | 4.08 | 0.3612 | 21949 | 6.1 | 1.05 |
| 423.83 | 0.959 | 3.99 | -3.17 | 0.254 | -0.647 | -- | 7.82 | -0.404 | 2.17 | 1.049 | 17035 | 5.03 | 0.933 |
| 482.03 | 1.02 | 3.81 | -1.59 | 0.22 | -1.69 | -- | -- | -0.00255 | 2.7 | 0.42 | 22032 | 6.36 | 0.7 |
| 427.84 | 0.963 | 3.98 | -3.17 | -- | -0.642 | -- | 7.82 | -0.399 | -- | 1.049 | 17016 | 5.03 | 0.923 |
| 510.13 | 1.06 | 3.67 | -0.0351 | 0.255 | -- | 0.448 | -- | 0.536 | 2.7 | -- | -- | -- | -- |
| 510.37 | 1.06 | 3.67 | -0.0467 | 0.279 | -- | -- | -- | 0.532 | 3.96 | -- | -- | -- | -- |
| 482.69 | 1.02 | 3.78 | -1.51 | -- | -1.53 | -- | -- | 0.0639 | -- | 0.3819 | 22019 | 6.37 | 0.418 |
| 498.82 | 1.04 | 3.74 | -- | 0.239 | -0.581 | -- | -- | 0.39 | 2.84 | 0.2522 | 16175 | 6.39 | 0.1 |
| 514.65 | 1.06 | 3.67 | -0.0353 | -- | -- | -- | -- | 0.539 | -- | -- | -- | -- | -- |
| 510.43 | 1.06 | 3.67 | -- | 0.279 | -- | -- | -- | 0.537 | 3.96 | -- | -- | -- | -- |
| 498.94 | 1.04 | 3.7 | -- | -- | -0.584 | -- | -- | 0.448 | -- | 0.00301 | 16528 | 6.51 | 0.0333 |
| 514.68 | 1.06 | 3.67 | -- | -- | -- | -- | -- | 0.543 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 416.18 | radiation | 10 | 43200 | > th | dose independent | rect. | 5 | 2 |
| 13 | 416.33 | wind | 1 | 43200 | < th | dose independent | rect. | 19 | 7 |
| 15 | 416.58 | humidity | 90 | 43200 | < th | dose independent | rect. | 20 | 4 |
| 16 | 416.88 | humidity | 90 | 43200 | < th | dose independent | rect. | 22 | 2 |
| 18 | 417.47 | wind | 1 | 43200 | < th | dose independent | rect. | 21 | 5 |
| 19 | 417.82 | wind | 1 | 43200 | < th | dose dependent | rect. | 16 | 10 |
| 22 | 418.49 | wind | 1 | 43200 | > th | dose independent | rect. | 19 | 7 |
| 23 | 418.89 | temperature | 30 | 43200 | > th | dose dependent | rect. | 14 | 1 |
| 26 | 419.02 | humidity | 90 | 43200 | > th | dose dependent | rect. | 5 | 1 |
| 30 | 419.23 | wind | 1 | 43200 | < th | dose dependent | rect. | 19 | 7 |
| 34 | 419.33 | wind | 1 | 43200 | > th | dose independent | rect. | 21 | 5 |
| 40 | 419.45 | temperature | 30 | 43200 | > th | dose dependent | rect. | 8 | 8 |
| 80 | 419.78 | temperature | 30 | 43200 | > th | dose dependent | sin | 4 | NA |
| 107 | 419.90 | temperature | 30 | 43200 | > th | dose independent | rect. | 8 | 8 |
| 136 | 419.96 | temperature | 30 | 43200 | > th | dose independent | rect. | 10 | 6 |
| 158 | 420.04 | temperature | 30 | 43200 | > th | dose independent | rect. | 12 | 4 |
| 178 | 420.07 | temperature | 30 | 43200 | > th | dose dependent | rect. | 11 | 22 |
| 180 | 420.08 | temperature | 30 | 43200 | > th | dose independent | rect. | 14 | 3 |
| 211 | 420.10 | temperature | 30 | 43200 | > th | dose independent | rect. | 14 | 1 |
| 213 | 420.10 | temperature | 30 | 43200 | < th | dose independent | rect. | 8 | 8 |
| 215 | 420.10 | temperature | 30 | 43200 | < th | dose independent | rect. | 4 | 12 |
| 217 | 420.10 | temperature | 30 | 43200 | < th | dose independent | rect. | 24 | 16 |
| 223 | 420.10 | temperature | 30 | 43200 | > th | dose dependent | no | NA | NA |
| 372 | 420.19 | temperature | 30 | 43200 | < th | dose independent | rect. | 10 | 6 |
| 382 | 420.27 | temperature | 30 | 43200 | > th | dose dependent | rect. | 14 | 23 |
| 387 | 420.31 | temperature | 30 | 43200 | > th | dose independent | sin | 2 | NA |
| 410 | 420.36 | temperature | 30 | 43200 | < th | dose independent | rect. | 12 | 5 |
| 445 | 420.40 | temperature | 30 | 43200 | > th | dose independent | rect. | 14 | 21 |
| 450 | 420.42 | temperature | 30 | 43200 | > th | dose independent | rect. | 12 | 23 |
| 452 | 420.43 | temperature | 30 | 43200 | > th | dose independent | rect. | 14 | 23 |
| 454 | 420.45 | temperature | 30 | 43200 | < th | dose independent | rect. | 14 | 3 |
| 458 | 420.46 | temperature | 30 | 43200 | < th | dose independent | sin | 2 | NA |
| 476 | 420.48 | temperature | 30 | 43200 | < th | dose independent | rect. | 14 | 21 |
| 482 | 420.50 | temperature | 30 | 43200 | > th | dose independent | rect. | 10 | 23 |
| 487 | 420.52 | humidity | 90 | 43200 | > th | dose independent | rect. | 20 | 4 |
| 488 | 420.52 | temperature | 30 | 43200 | < th | dose independent | rect. | 12 | 23 |
| 489 | 420.52 | temperature | 30 | 43200 | < th | dose independent | rect. | 14 | 23 |
| 491 | 420.53 | temperature | 30 | 43200 | > th | dose independent | no | NA | NA |
| 614 | 420.60 | temperature | 30 | 43200 | < th | dose independent | rect. | 10 | 23 |
| 656 | 420.63 | temperature | 30 | 43200 | < th | dose independent | rect. | 8 | 17 |
| 663 | 420.63 | temperature | 30 | 43200 | < th | dose independent | rect. | 5 | 20 |
| 670 | 420.63 | temperature | 30 | 43200 | < th | dose independent | rect. | 8 | 22 |
| 674 | 420.64 | temperature | 30 | 43200 | < th | dose independent | no | NA | NA |
| 769 | 420.72 | temperature | 30 | 43200 | < th | dose independent | rect. | 10 | 15 |
| 771 | 420.72 | temperature | 30 | 43200 | < th | dose independent | rect. | 10 | 20 |
| 859 | 420.95 | humidity | 90 | 43200 | > th | dose dependent | rect. | 1 | 9 |
| 860 | 420.96 | temperature | 30 | 43200 | < th | dose independent | rect. | 12 | 13 |
| 863 | 420.97 | temperature | 30 | 43200 | < th | dose independent | rect. | 12 | 11 |
| 964 | 421.21 | wind | 1 | 43200 | < th | dose dependent | rect. | 21 | 5 |
| 966 | 421.22 | temperature | 30 | 43200 | > th | dose independent | rect. | 10 | 1 |
| 969 | 421.24 | temperature | 30 | 43200 | < th | dose independent | rect. | 10 | 1 |
| 990 | 421.27 | humidity | 90 | 43200 | > th | dose dependent | rect. | 4 | 3 |