Os05g0104900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Clone ZZD1296 mRNA sequence.


FiT-DB / Search/ Help/ Sample detail
|
Os05g0104900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Clone ZZD1296 mRNA sequence.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
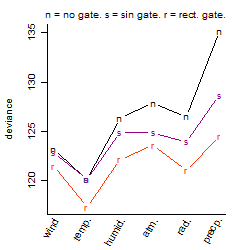
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 119.53 | 0.514 | 9.77 | -2.57 | 0.162 | -0.894 | -0.3 | 2.57 | -0.399 | 21.9 | 26.7 | 1361 | -- | -- |
| 124.46 | 0.52 | 9.69 | -1.88 | 0.154 | -0.687 | -0.436 | -- | -0.234 | 22.1 | 26.3 | 1431 | -- | -- |
| 119.84 | 0.51 | 9.77 | -2.58 | 0.168 | -0.886 | -- | 2.62 | -0.403 | 22.4 | 26.7 | 1374 | -- | -- |
| 124.69 | 0.52 | 9.68 | -1.79 | 0.176 | -0.667 | -- | -- | -0.206 | 22 | 26.6 | 1401 | -- | -- |
| 121.48 | 0.513 | 9.77 | -2.57 | -- | -0.913 | -- | 2.64 | -0.407 | -- | 26.71 | 1357 | -- | -- |
| 140.27 | 0.555 | 9.64 | -1.56 | 0.206 | -- | -0.278 | -- | 0.00173 | 23 | -- | -- | -- | -- |
| 140.58 | 0.555 | 9.64 | -1.55 | 0.207 | -- | -- | -- | 0.00227 | 23.3 | -- | -- | -- | -- |
| 126.16 | 0.523 | 9.68 | -1.8 | -- | -0.704 | -- | -- | -0.182 | -- | 26.66 | 1327 | -- | -- |
| 199.82 | 0.658 | 9.62 | -- | 0.15 | -0.296 | -- | -- | 0.0893 | 21.7 | 22.4 | 666 | -- | -- |
| 143.27 | 0.559 | 9.63 | -1.55 | -- | -- | -- | -- | 0.00423 | -- | -- | -- | -- | -- |
| 204.15 | 0.668 | 9.6 | -- | 0.193 | -- | -- | -- | 0.175 | 22.8 | -- | -- | -- | -- |
| 200.84 | 0.66 | 9.62 | -- | -- | -0.327 | -- | -- | 0.0743 | -- | 22.3 | 688 | -- | -- |
| 206.46 | 0.671 | 9.6 | -- | -- | -- | -- | -- | 0.176 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 117.24 | temperature | 15 | 270 | > th | dose dependent | rect. | 14 | 17 |
| 2 | 117.32 | temperature | 15 | 270 | > th | dose dependent | rect. | 12 | 21 |
| 20 | 118.43 | temperature | 10 | 270 | > th | dose dependent | rect. | 3 | 16 |
| 24 | 118.59 | temperature | 10 | 270 | > th | dose dependent | rect. | 24 | 22 |
| 36 | 118.98 | temperature | 25 | 1440 | > th | dose dependent | rect. | 16 | 2 |
| 69 | 119.50 | temperature | 30 | 270 | < th | dose dependent | rect. | 7 | 20 |
| 95 | 119.69 | temperature | 20 | 720 | > th | dose dependent | rect. | 19 | 23 |
| 97 | 119.72 | temperature | 25 | 1440 | > th | dose dependent | rect. | 11 | 8 |
| 100 | 119.76 | temperature | 25 | 1440 | < th | dose independent | rect. | 17 | 5 |
| 105 | 119.78 | temperature | 20 | 720 | > th | dose dependent | rect. | 19 | 20 |
| 119 | 119.84 | temperature | 25 | 1440 | > th | dose independent | rect. | 17 | 5 |
| 128 | 119.89 | temperature | 20 | 1440 | > th | dose dependent | rect. | 15 | 8 |
| 171 | 120.02 | temperature | 20 | 1440 | > th | dose dependent | rect. | 20 | 1 |
| 173 | 120.02 | temperature | 20 | 10 | > th | dose dependent | rect. | 16 | 16 |
| 208 | 120.09 | temperature | 20 | 1440 | > th | dose dependent | sin | 15 | NA |
| 216 | 120.11 | temperature | 20 | 720 | > th | dose dependent | no | NA | NA |
| 240 | 120.14 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 23 |
| 390 | 120.37 | temperature | 20 | 720 | > th | dose dependent | sin | 11 | NA |
| 448 | 120.43 | temperature | 25 | 1440 | > th | dose independent | rect. | 19 | 1 |
| 575 | 120.58 | temperature | 30 | 720 | < th | dose dependent | rect. | 15 | 20 |
| 867 | 120.94 | temperature | 25 | 1440 | > th | dose dependent | rect. | 12 | 1 |
| 929 | 121.01 | radiation | 1 | 14400 | > th | dose independent | rect. | 5 | 1 |