Os03g0243700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Glycoside hydrolase, family 10 protein.

FiT-DB / Search/ Help/ Sample detail
|
Os03g0243700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Glycoside hydrolase, family 10 protein.
|
|
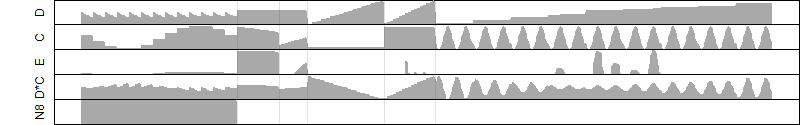
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

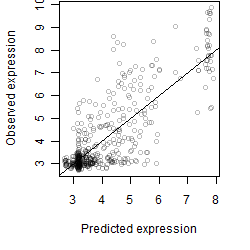 __
__
Dependence on each variable
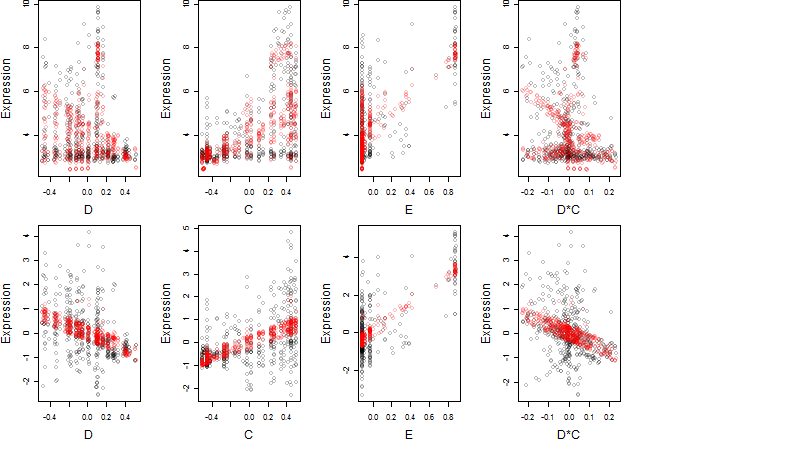
Residual plot
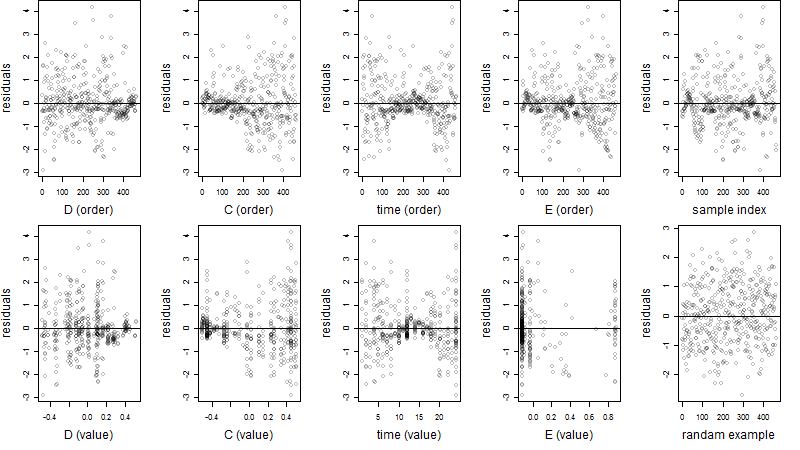
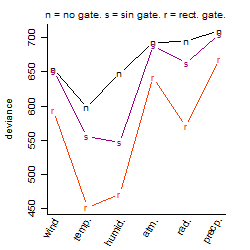
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 435.57 | 0.981 | 4.22 | -0.903 | 1.31 | 3.18 | -3.67 | 6.15 | -0.514 | 1.9 | 25.88 | 671 | 18.5 | 4.73 |
| 437.45 | 0.974 | 4.23 | -1.53 | 1.42 | 3.79 | -3.6 | -- | -0.367 | 1.84 | 26.3 | 675 | 18.6 | 5.13 |
| 485.89 | 1.03 | 4.19 | -0.868 | 1.44 | 3.05 | -- | 5.69 | -0.434 | 2.09 | 26.03 | 667 | 18.5 | 4.72 |
| 483.25 | 1.02 | 4.26 | -1.48 | 1.41 | 3.65 | -- | -- | -0.491 | 2.2 | 26.12 | 659 | 18.6 | 4.83 |
| 557.2 | 1.1 | 4.3 | -0.908 | -- | 3.92 | -- | 4.65 | -0.817 | -- | 24.64 | 652 | 18.5 | 4.85 |
| 831.14 | 1.35 | 4.32 | -1.05 | 2.57 | -- | -1.87 | -- | -0.76 | 2.67 | -- | -- | -- | -- |
| 843.05 | 1.36 | 4.31 | -1 | 2.58 | -- | -- | -- | -0.75 | 2.8 | -- | -- | -- | -- |
| 553.53 | 1.1 | 4.35 | -1.53 | -- | 4.42 | -- | -- | -0.884 | -- | 25.07 | 659 | 18.5 | 5.24 |
| 528.56 | 1.07 | 4.23 | -- | 1.51 | 3.44 | -- | -- | -0.358 | 1.87 | 26.15 | 581 | 20 | 2.47 |
| 1228.35 | 1.64 | 4.3 | -0.89 | -- | -- | -- | -- | -0.684 | -- | -- | -- | -- | -- |
| 869.42 | 1.38 | 4.29 | -- | 2.56 | -- | -- | -- | -0.639 | 2.8 | -- | -- | -- | -- |
| 615.2 | 1.16 | 4.29 | -- | -- | 4.26 | -- | -- | -0.627 | -- | 25.41 | 581 | 19.9 | 2.74 |
| 1249.26 | 1.65 | 4.28 | -- | -- | -- | -- | -- | -0.585 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance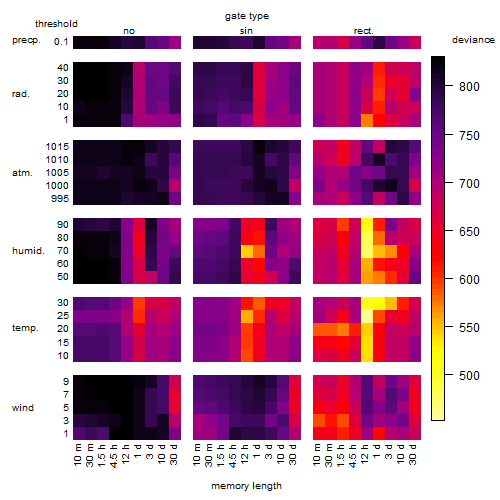
|
Histogram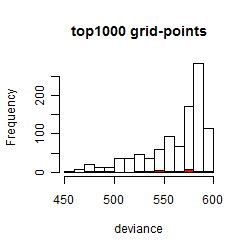 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 452.32 | temperature | 25 | 720 | > th | dose dependent | rect. | 18 | 5 |
| 12 | 471.00 | humidity | 70 | 720 | < th | dose independent | rect. | 18 | 2 |
| 33 | 481.19 | humidity | 80 | 720 | < th | dose dependent | rect. | 18 | 3 |
| 49 | 495.91 | humidity | 70 | 720 | > th | dose independent | rect. | 17 | 8 |
| 145 | 528.29 | temperature | 30 | 720 | > th | dose independent | rect. | 18 | 1 |
| 179 | 534.35 | humidity | 70 | 720 | > th | dose independent | rect. | 17 | 3 |
| 183 | 535.65 | temperature | 15 | 720 | > th | dose dependent | rect. | 17 | 6 |
| 225 | 545.55 | humidity | 70 | 720 | > th | dose independent | rect. | 17 | 16 |
| 228 | 546.19 | humidity | 70 | 720 | > th | dose independent | sin | 12 | NA |
| 233 | 547.09 | temperature | 30 | 720 | < th | dose independent | rect. | 17 | 8 |
| 259 | 548.38 | humidity | 70 | 1440 | > th | dose independent | rect. | 18 | 13 |
| 335 | 553.49 | humidity | 60 | 720 | > th | dose dependent | rect. | 17 | 6 |
| 344 | 555.65 | temperature | 25 | 720 | > th | dose dependent | sin | 12 | NA |
| 364 | 560.47 | humidity | 70 | 720 | < th | dose independent | sin | 12 | NA |
| 384 | 564.64 | humidity | 70 | 720 | > th | dose dependent | rect. | 19 | 2 |
| 419 | 568.83 | temperature | 30 | 720 | < th | dose dependent | rect. | 17 | 6 |
| 433 | 570.66 | radiation | 1 | 720 | > th | dose independent | rect. | 18 | 11 |
| 435 | 570.71 | radiation | 1 | 720 | > th | dose independent | rect. | 18 | 1 |
| 488 | 571.87 | temperature | 30 | 1440 | < th | dose independent | rect. | 17 | 15 |
| 538 | 574.62 | humidity | 50 | 1440 | < th | dose independent | rect. | 16 | 1 |
| 543 | 575.81 | temperature | 20 | 90 | > th | dose dependent | rect. | 3 | 2 |
| 546 | 576.32 | humidity | 50 | 1440 | < th | dose dependent | rect. | 16 | 1 |
| 553 | 576.94 | temperature | 30 | 720 | < th | dose dependent | rect. | 18 | 3 |
| 659 | 582.94 | temperature | 20 | 10 | > th | dose dependent | rect. | 3 | 3 |
| 708 | 583.96 | temperature | 30 | 1440 | < th | dose independent | sin | 12 | NA |
| 725 | 585.07 | temperature | 30 | 1440 | > th | dose independent | sin | 12 | NA |
| 994 | 593.45 | wind | 3 | 90 | < th | dose dependent | rect. | 3 | 2 |
| 995 | 593.45 | wind | 3 | 10 | < th | dose dependent | rect. | 3 | 3 |