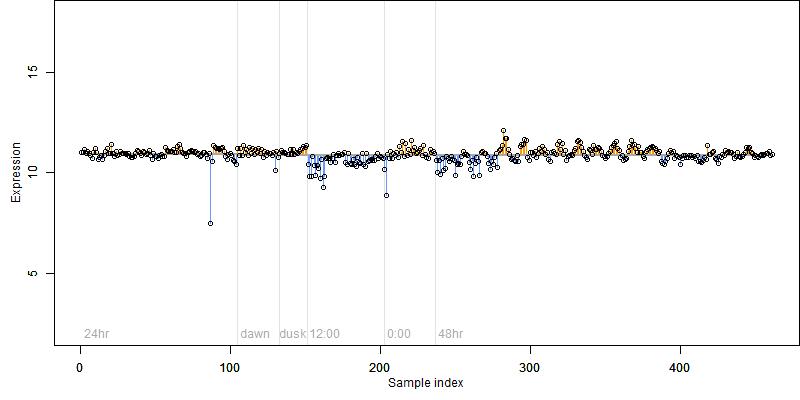
Os03g0225600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved oligomeric complex COG6 family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os03g0225600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved oligomeric complex COG6 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 41.03 | 0.301 | 10.9 | -0.0282 | 0.347 | -0.706 | 0.0975 | 1.09 | -0.088 | 0.305 | 23.4 | 1185 | -- | -- |
| 42.55 | 0.304 | 10.9 | 0.0625 | 0.362 | -0.691 | -0.0325 | -- | -0.0685 | 0.312 | 22.74 | 2594 | -- | -- |
| 41.06 | 0.298 | 10.9 | -0.0231 | 0.348 | -0.715 | -- | 1.07 | -0.0848 | 0.284 | 23.26 | 1213 | -- | -- |
| 42.26 | 0.303 | 10.9 | 0.011 | 0.356 | -0.888 | -- | -- | -0.0585 | 0.232 | 20.88 | 2582 | -- | -- |
| 47.26 | 0.32 | 11 | -0.164 | -- | -0.863 | -- | 1.52 | -0.141 | -- | 31.67 | 950 | -- | -- |
| 51.41 | 0.336 | 10.9 | 0.366 | 0.397 | -- | -0.0621 | -- | 0.0889 | 0.463 | -- | -- | -- | -- |
| 51.43 | 0.336 | 10.9 | 0.368 | 0.397 | -- | -- | -- | 0.0892 | 0.445 | -- | -- | -- | -- |
| 49.15 | 0.327 | 10.9 | 0.124 | -- | -0.852 | -- | -- | -0.0976 | -- | 26.79 | 957 | -- | -- |
| 42.26 | 0.303 | 10.9 | -- | 0.356 | -0.897 | -- | -- | -0.0608 | 0.227 | 20.89 | 2583 | -- | -- |
| 61.33 | 0.366 | 10.9 | 0.382 | -- | -- | -- | -- | 0.0958 | -- | -- | -- | -- | -- |
| 55 | 0.347 | 10.9 | -- | 0.403 | -- | -- | -- | 0.0482 | 0.492 | -- | -- | -- | -- |
| 49.41 | 0.327 | 10.9 | -- | -- | -0.927 | -- | -- | -0.125 | -- | 26.79 | 958 | -- | -- |
| 65.18 | 0.377 | 10.9 | -- | -- | -- | -- | -- | 0.0534 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance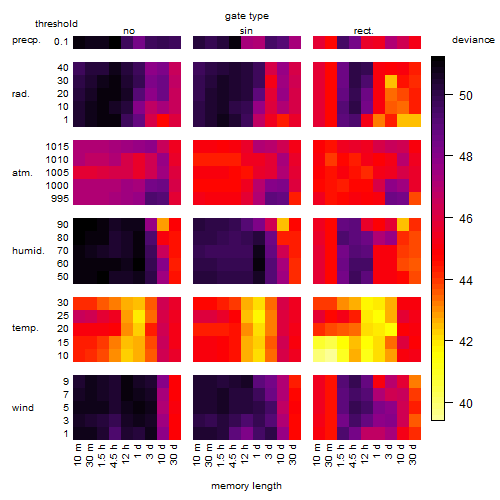
|
Histogram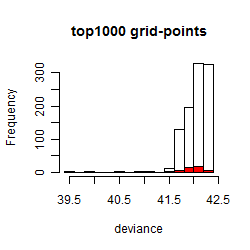 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 39.40 | temperature | 10 | 30 | > th | dose dependent | rect. | 3 | 23 |
| 6 | 41.07 | temperature | 10 | 720 | > th | dose dependent | rect. | 18 | 21 |
| 13 | 41.58 | temperature | 20 | 4320 | < th | dose independent | rect. | 19 | 3 |
| 14 | 41.58 | temperature | 25 | 1440 | < th | dose dependent | rect. | 17 | 18 |
| 15 | 41.59 | temperature | 25 | 1440 | < th | dose dependent | rect. | 17 | 11 |
| 31 | 41.65 | temperature | 25 | 1440 | < th | dose dependent | sin | 13 | NA |
| 65 | 41.71 | temperature | 20 | 4320 | > th | dose independent | rect. | 19 | 3 |
| 69 | 41.71 | temperature | 15 | 30 | > th | dose independent | rect. | 3 | 23 |
| 70 | 41.71 | temperature | 25 | 1440 | < th | dose dependent | rect. | 19 | 5 |
| 92 | 41.73 | temperature | 25 | 1440 | < th | dose dependent | rect. | 22 | 1 |
| 96 | 41.73 | temperature | 30 | 720 | < th | dose dependent | rect. | 1 | 18 |
| 149 | 41.81 | temperature | 20 | 4320 | < th | dose independent | rect. | 21 | 1 |
| 162 | 41.82 | temperature | 25 | 1440 | < th | dose dependent | no | NA | NA |
| 166 | 41.82 | temperature | 25 | 1440 | < th | dose dependent | rect. | 8 | 22 |
| 196 | 41.85 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 23 |
| 210 | 41.86 | temperature | 25 | 1440 | < th | dose dependent | rect. | 3 | 23 |
| 221 | 41.88 | temperature | 10 | 720 | > th | dose dependent | rect. | 3 | 19 |
| 227 | 41.89 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 10 |
| 239 | 41.89 | temperature | 10 | 720 | > th | dose dependent | rect. | 3 | 21 |
| 247 | 41.90 | temperature | 30 | 1440 | < th | dose dependent | rect. | 3 | 1 |
| 287 | 41.95 | temperature | 25 | 720 | < th | dose dependent | rect. | 4 | 18 |
| 291 | 41.96 | temperature | 20 | 1440 | > th | dose independent | rect. | 12 | 19 |
| 320 | 41.98 | temperature | 20 | 1440 | < th | dose independent | rect. | 12 | 18 |
| 339 | 41.99 | temperature | 10 | 720 | > th | dose dependent | rect. | 12 | 23 |
| 381 | 42.05 | temperature | 10 | 720 | > th | dose dependent | rect. | 1 | 23 |
| 415 | 42.08 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 17 |
| 419 | 42.09 | temperature | 10 | 720 | > th | dose dependent | rect. | 3 | 12 |
| 457 | 42.11 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 13 |
| 466 | 42.12 | temperature | 15 | 1440 | > th | dose dependent | rect. | 3 | 1 |
| 484 | 42.13 | temperature | 20 | 1440 | > th | dose independent | rect. | 15 | 16 |
| 488 | 42.13 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 4 |
| 506 | 42.14 | temperature | 20 | 1440 | > th | dose independent | rect. | 17 | 14 |
| 509 | 42.14 | temperature | 10 | 1440 | > th | dose dependent | sin | 10 | NA |
| 527 | 42.15 | temperature | 20 | 1440 | > th | dose independent | sin | 13 | NA |
| 529 | 42.15 | temperature | 20 | 1440 | < th | dose independent | rect. | 17 | 13 |
| 554 | 42.16 | temperature | 10 | 720 | > th | dose dependent | rect. | 23 | 16 |
| 567 | 42.17 | temperature | 20 | 1440 | < th | dose independent | rect. | 15 | 15 |
| 586 | 42.18 | temperature | 20 | 1440 | < th | dose independent | rect. | 21 | 18 |
| 590 | 42.18 | temperature | 10 | 1440 | > th | dose dependent | rect. | 1 | 9 |
| 645 | 42.20 | temperature | 20 | 1440 | < th | dose independent | sin | 13 | NA |
| 649 | 42.20 | temperature | 20 | 1440 | > th | dose independent | rect. | 21 | 18 |
| 722 | 42.22 | temperature | 10 | 1440 | > th | dose dependent | rect. | 1 | 4 |
| 753 | 42.23 | temperature | 10 | 720 | > th | dose dependent | rect. | 1 | 14 |
| 769 | 42.24 | temperature | 25 | 1440 | < th | dose dependent | rect. | 1 | 1 |
| 770 | 42.24 | temperature | 25 | 1440 | < th | dose dependent | rect. | 8 | 1 |
| 846 | 42.26 | temperature | 20 | 1440 | > th | dose independent | rect. | 18 | 9 |
| 962 | 42.29 | temperature | 25 | 1440 | < th | dose dependent | rect. | 7 | 3 |