Os02g0589000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Lecithin:cholesterol acyltransferase family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os02g0589000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Lecithin:cholesterol acyltransferase family protein.
|
|
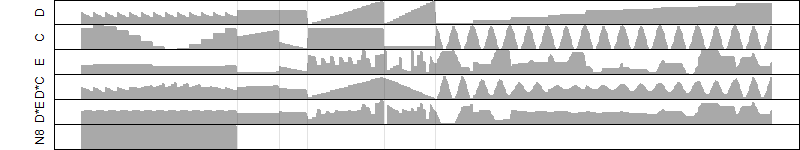
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 536.86 | 1.09 | 13 | -5.33 | 2.07 | -4.47 | 4.63 | -4.59 | -0.574 | 9.54 | 75 | 1300 | -- | -- |
| 578.79 | 1.12 | 13 | -6.02 | 2.17 | -4.7 | 4.3 | -- | -0.757 | 9.53 | 74.81 | 1275 | -- | -- |
| 614.61 | 1.15 | 13 | -5.22 | 1.97 | -4.39 | -- | -4.08 | -0.583 | 9.43 | 74.89 | 1317 | -- | -- |
| 646.35 | 1.18 | 13 | -5.84 | 2.14 | -4.64 | -- | -- | -0.751 | 9.63 | 74.83 | 1275 | -- | -- |
| 782.7 | 1.3 | 13 | -5.11 | -- | -4.13 | -- | -4.51 | -0.518 | -- | 74.81 | 1448 | -- | -- |
| 1231.58 | 1.65 | 12.9 | -5.26 | 1.83 | -- | 4.22 | -- | -0.142 | 8.2 | -- | -- | -- | -- |
| 1285.88 | 1.68 | 12.9 | -5.13 | 1.83 | -- | -- | -- | -0.145 | 8.11 | -- | -- | -- | -- |
| 830 | 1.34 | 13.1 | -5.79 | -- | -4.37 | -- | -- | -0.703 | -- | 74.25 | 1429 | -- | -- |
| 1270.99 | 1.66 | 13 | -- | 3.49 | -6.54 | -- | -- | -0.254 | 8.55 | 82.04 | 2185 | -- | -- |
| 1468.84 | 1.79 | 12.9 | -5.12 | -- | -- | -- | -- | -0.123 | -- | -- | -- | -- | -- |
| 1980.74 | 2.08 | 12.8 | -- | 1.8 | -- | -- | -- | 0.427 | 8.31 | -- | -- | -- | -- |
| 1528.58 | 1.82 | 13 | -- | -- | -6.62 | -- | -- | -0.297 | -- | 85 | 1518 | -- | -- |
| 2160.01 | 2.17 | 12.8 | -- | -- | -- | -- | -- | 0.445 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 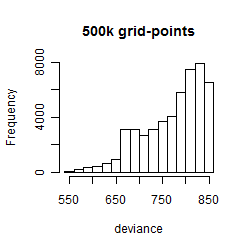
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 548.25 | humidity | 80 | 1440 | < th | dose independent | rect. | 7 | 14 |
| 2 | 548.63 | humidity | 70 | 1440 | > th | dose dependent | rect. | 6 | 16 |
| 22 | 555.67 | humidity | 80 | 1440 | > th | dose independent | rect. | 6 | 15 |
| 153 | 573.74 | humidity | 60 | 1440 | > th | dose dependent | sin | 22 | NA |
| 179 | 575.78 | humidity | 50 | 1440 | > th | dose dependent | rect. | 13 | 23 |
| 181 | 576.00 | humidity | 50 | 1440 | > th | dose dependent | rect. | 16 | 23 |
| 184 | 576.22 | humidity | 60 | 1440 | > th | dose dependent | no | NA | NA |
| 185 | 576.27 | humidity | 50 | 1440 | > th | dose dependent | rect. | 10 | 23 |
| 204 | 577.55 | humidity | 80 | 1440 | < th | dose independent | sin | 0 | NA |
| 300 | 585.19 | humidity | 80 | 1440 | > th | dose independent | sin | 0 | NA |
| 356 | 589.20 | humidity | 90 | 1440 | < th | dose dependent | rect. | 16 | 22 |
| 416 | 593.38 | humidity | 90 | 1440 | < th | dose dependent | rect. | 5 | 18 |
| 483 | 596.51 | humidity | 90 | 1440 | < th | dose dependent | rect. | 10 | 23 |
| 493 | 597.01 | humidity | 80 | 1440 | < th | dose independent | rect. | 7 | 9 |
| 511 | 597.59 | humidity | 90 | 1440 | < th | dose dependent | no | NA | NA |
| 576 | 601.87 | humidity | 70 | 1440 | > th | dose dependent | rect. | 6 | 10 |
| 676 | 608.31 | humidity | 50 | 1440 | > th | dose dependent | rect. | 13 | 13 |
| 708 | 609.97 | humidity | 90 | 1440 | < th | dose dependent | sin | 17 | NA |
| 756 | 611.80 | humidity | 80 | 1440 | > th | dose independent | rect. | 7 | 9 |
| 778 | 612.94 | humidity | 50 | 1440 | > th | dose dependent | rect. | 15 | 8 |
| 788 | 613.25 | humidity | 70 | 1440 | > th | dose independent | rect. | 18 | 23 |
| 800 | 613.62 | humidity | 70 | 14400 | < th | dose independent | rect. | 18 | 13 |
| 811 | 614.33 | humidity | 70 | 14400 | < th | dose independent | rect. | 18 | 2 |
| 814 | 614.43 | humidity | 70 | 1440 | > th | dose independent | rect. | 13 | 21 |
| 821 | 614.79 | humidity | 70 | 1440 | > th | dose independent | rect. | 13 | 23 |
| 826 | 615.05 | humidity | 70 | 14400 | < th | dose independent | rect. | 18 | 4 |
| 876 | 617.17 | humidity | 70 | 1440 | > th | dose independent | rect. | 11 | 23 |
| 946 | 619.54 | humidity | 70 | 1440 | > th | dose independent | sin | 19 | NA |
| 984 | 620.17 | humidity | 70 | 1440 | > th | dose independent | no | NA | NA |