Os02g0558600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.
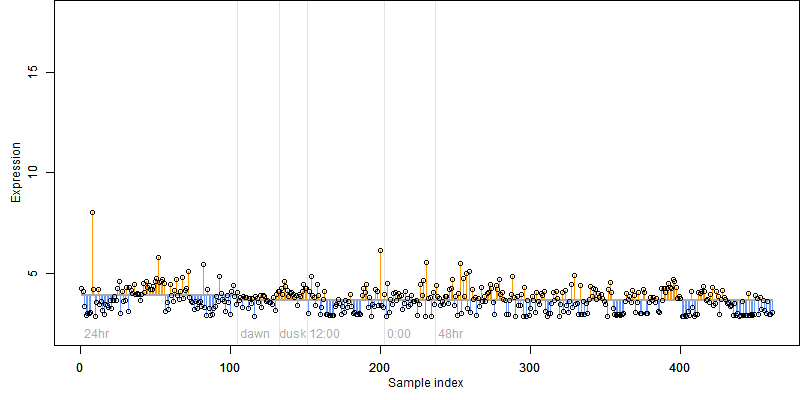

FiT-DB / Search/ Help/ Sample detail
|
Os02g0558600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable

Residual plot

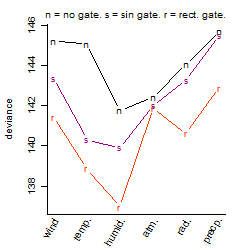
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 141.68 | 0.559 | 3.59 | -0.447 | 0.396 | 0.516 | 0.0603 | 1.73 | 0.351 | 19.8 | 81 | 4325 | -- | -- |
| 143.57 | 0.558 | 3.59 | -0.281 | 0.479 | 0.651 | 0.0605 | -- | 0.472 | 19.5 | 82 | 4747 | -- | -- |
| 141.69 | 0.554 | 3.59 | -0.45 | 0.396 | 0.515 | -- | 1.74 | 0.349 | 19.8 | 81 | 4318 | -- | -- |
| 143.56 | 0.558 | 3.59 | -0.284 | 0.464 | 0.659 | -- | -- | 0.474 | 19.4 | 81.5 | 4731 | -- | -- |
| 150.09 | 0.571 | 3.58 | -0.477 | -- | 0.494 | -- | 1.8 | 0.339 | -- | 82 | 4291 | -- | -- |
| 148.38 | 0.571 | 3.64 | -0.239 | 0.397 | -- | 0.0748 | -- | 0.212 | 19.9 | -- | -- | -- | -- |
| 148.4 | 0.57 | 3.64 | -0.241 | 0.397 | -- | -- | -- | 0.212 | 19.9 | -- | -- | -- | -- |
| 152.82 | 0.576 | 3.58 | -0.271 | -- | 0.549 | -- | -- | 0.438 | -- | 77.83 | 5779 | -- | -- |
| 145.7 | 0.562 | 3.58 | -- | 0.45 | 0.624 | -- | -- | 0.492 | 19.4 | 81.44 | 4693 | -- | -- |
| 156.95 | 0.585 | 3.63 | -0.246 | -- | -- | -- | -- | 0.207 | -- | -- | -- | -- | -- |
| 149.93 | 0.573 | 3.64 | -- | 0.398 | -- | -- | -- | 0.239 | 19.8 | -- | -- | -- | -- |
| 154.62 | 0.579 | 3.57 | -- | -- | 0.515 | -- | -- | 0.466 | -- | 82 | 5764 | -- | -- |
| 158.54 | 0.588 | 3.63 | -- | -- | -- | -- | -- | 0.234 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance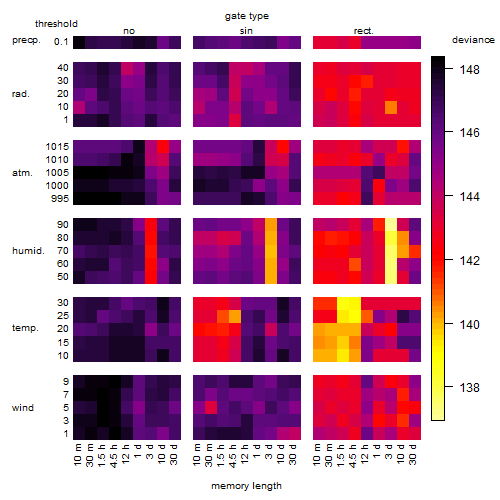
|
Histogram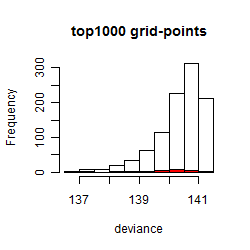 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 136.94 | humidity | 90 | 4320 | < th | dose dependent | rect. | 5 | 3 |
| 3 | 137.35 | humidity | 60 | 4320 | > th | dose dependent | rect. | 4 | 5 |
| 6 | 137.45 | humidity | 90 | 4320 | < th | dose dependent | rect. | 4 | 5 |
| 19 | 138.12 | humidity | 80 | 4320 | > th | dose independent | rect. | 5 | 4 |
| 44 | 138.67 | humidity | 80 | 4320 | < th | dose independent | rect. | 5 | 4 |
| 58 | 138.86 | temperature | 30 | 90 | < th | dose dependent | rect. | 3 | 8 |
| 64 | 138.93 | temperature | 25 | 270 | < th | dose independent | rect. | 23 | 10 |
| 92 | 139.15 | temperature | 30 | 270 | < th | dose dependent | rect. | 1 | 8 |
| 118 | 139.41 | temperature | 25 | 90 | > th | dose independent | rect. | 23 | 15 |
| 122 | 139.42 | temperature | 10 | 90 | > th | dose dependent | rect. | 23 | 15 |
| 186 | 139.75 | humidity | 80 | 4320 | > th | dose independent | rect. | 23 | 11 |
| 194 | 139.77 | temperature | 25 | 90 | < th | dose independent | rect. | 3 | 8 |
| 198 | 139.78 | temperature | 25 | 270 | > th | dose independent | rect. | 20 | 16 |
| 228 | 139.91 | humidity | 70 | 4320 | > th | dose dependent | sin | 5 | NA |
| 257 | 140.02 | temperature | 20 | 270 | > th | dose dependent | rect. | 20 | 16 |
| 285 | 140.10 | humidity | 80 | 4320 | > th | dose independent | sin | 5 | NA |
| 319 | 140.17 | temperature | 10 | 270 | > th | dose dependent | rect. | 21 | 15 |
| 334 | 140.20 | temperature | 20 | 90 | > th | dose dependent | rect. | 20 | 18 |
| 370 | 140.30 | temperature | 25 | 270 | < th | dose independent | sin | 8 | NA |
| 380 | 140.33 | temperature | 25 | 270 | > th | dose independent | sin | 8 | NA |
| 384 | 140.33 | humidity | 90 | 4320 | < th | dose dependent | sin | 7 | NA |
| 387 | 140.34 | humidity | 80 | 4320 | < th | dose independent | sin | 5 | NA |
| 537 | 140.62 | radiation | 10 | 4320 | < th | dose independent | rect. | 8 | 1 |
| 644 | 140.79 | humidity | 80 | 4320 | < th | dose independent | rect. | 21 | 16 |
| 692 | 140.85 | radiation | 10 | 4320 | < th | dose dependent | rect. | 8 | 1 |
| 787 | 141.00 | temperature | 25 | 720 | < th | dose dependent | rect. | 12 | 1 |
| 904 | 141.15 | humidity | 60 | 270 | > th | dose independent | rect. | 8 | 17 |
| 920 | 141.18 | temperature | 25 | 90 | > th | dose independent | sin | 5 | NA |