Os02g0543200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Cyclin-like F-box domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os02g0543200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Cyclin-like F-box domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

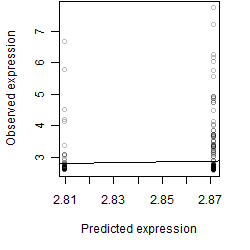 __
__
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 167.29 | 0.608 | 2.89 | -0.102 | 0.012 | 3.79 | 0.228 | 6.32 | -0.0477 | 14.9 | 9.1 | 26 | -- | -- |
| 168.93 | 0.605 | 2.87 | -0.143 | 0.0297 | 2.11 | 0.265 | -- | -0.06 | 14.4 | 9 | 28 | -- | -- |
| 167.43 | 0.603 | 2.89 | -0.0968 | 0.0224 | 3.83 | -- | 6.66 | -0.0475 | 11.2 | 9 | 25 | -- | -- |
| 169.27 | 0.606 | 2.87 | -0.143 | 0.0256 | 2.02 | -- | -- | -0.046 | 10.6 | 9.1 | 28 | -- | -- |
| 167.46 | 0.603 | 2.89 | -0.0973 | -- | 3.85 | -- | 6.68 | -0.0477 | -- | 9.007 | 25 | -- | -- |
| 179.45 | 0.628 | 2.88 | -0.263 | 0.0772 | -- | 0.0917 | -- | -0.0897 | 13.6 | -- | -- | -- | -- |
| 179.48 | 0.627 | 2.88 | -0.26 | 0.0782 | -- | -- | -- | -0.0891 | 14 | -- | -- | -- | -- |
| 166.11 | 0.6 | 2.86 | -0.0319 | -- | 3.84 | -- | -- | 0.0202 | -- | 10.6 | 28 | -- | -- |
| 169.73 | 0.607 | 2.86 | -- | 0.0254 | 2.13 | -- | -- | -0.0272 | 10.5 | 9.1 | 27 | -- | -- |
| 179.85 | 0.627 | 2.88 | -0.263 | -- | -- | -- | -- | -0.0909 | -- | -- | -- | -- | -- |
| 181.26 | 0.63 | 2.87 | -- | 0.0831 | -- | -- | -- | -0.0601 | 14.1 | -- | -- | -- | -- |
| 164.43 | 0.597 | 2.86 | -- | -- | 2.94 | -- | -- | -0.0421 | -- | 10.54 | 26 | -- | -- |
| 181.68 | 0.629 | 2.87 | -- | -- | -- | -- | -- | -0.0617 | -- | -- | -- | -- | -- |
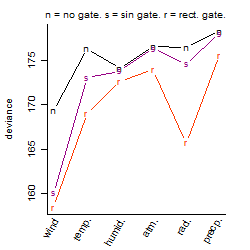
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 158.43 | wind | 9 | 30 | > th | dose dependent | rect. | 21 | 15 |
| 9 | 159.98 | wind | 9 | 30 | > th | dose dependent | rect. | 11 | 1 |
| 11 | 160.05 | wind | 9 | 30 | > th | dose dependent | sin | 7 | NA |
| 42 | 161.70 | wind | 9 | 30 | > th | dose independent | rect. | 21 | 15 |
| 48 | 162.13 | wind | 9 | 30 | < th | dose independent | sin | 7 | NA |
| 61 | 162.78 | wind | 9 | 30 | > th | dose independent | rect. | 11 | 1 |
| 86 | 163.72 | wind | 9 | 30 | > th | dose independent | sin | 7 | NA |
| 148 | 165.69 | radiation | 10 | 10 | < th | dose independent | rect. | 12 | 1 |
| 349 | 168.77 | wind | 9 | 30 | > th | dose independent | rect. | 11 | 11 |
| 370 | 169.03 | temperature | 30 | 90 | > th | dose dependent | rect. | 10 | 1 |
| 439 | 169.36 | wind | 9 | 30 | > th | dose independent | no | NA | NA |
| 486 | 169.43 | wind | 9 | 30 | < th | dose independent | no | NA | NA |
| 575 | 171.29 | temperature | 30 | 90 | > th | dose independent | rect. | 10 | 1 |
| 576 | 171.37 | wind | 1 | 30 | < th | dose independent | rect. | 22 | 2 |
| 605 | 171.67 | wind | 9 | 30 | > th | dose dependent | rect. | 11 | 11 |
| 671 | 171.83 | wind | 9 | 30 | > th | dose dependent | no | NA | NA |
| 845 | 172.68 | humidity | 60 | 4320 | > th | dose independent | rect. | 14 | 1 |
| 846 | 172.69 | wind | 3 | 270 | > th | dose dependent | rect. | 21 | 1 |
| 944 | 173.04 | wind | 5 | 270 | > th | dose independent | rect. | 21 | 1 |
| 947 | 173.06 | humidity | 60 | 4320 | < th | dose independent | rect. | 14 | 1 |
| 948 | 173.06 | temperature | 30 | 90 | > th | dose dependent | sin | 9 | NA |