Os02g0318100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Alternative oxidase 1a, mitochondrial precursor (EC 1.-.-.-).
FiT-DB / Search/ Help/ Sample detail
|
Os02g0318100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Alternative oxidase 1a, mitochondrial precursor (EC 1.-.-.-).
|
|
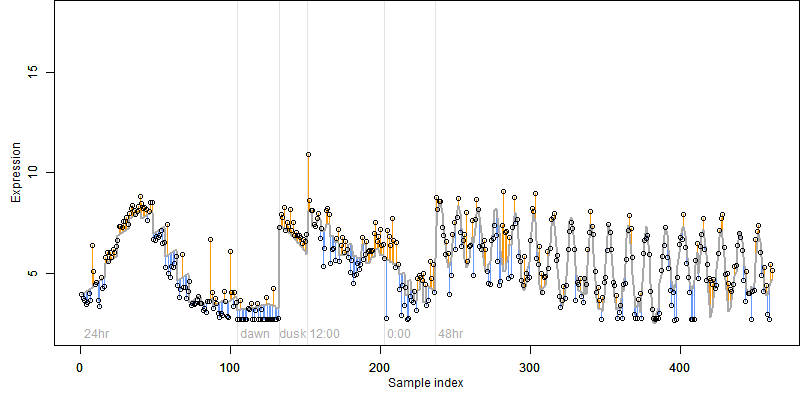
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
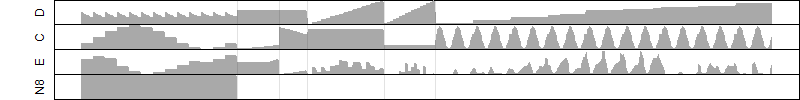
Dependence on each variable

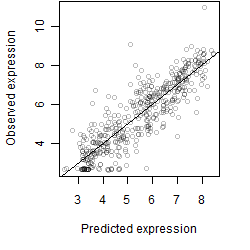
Residual plot

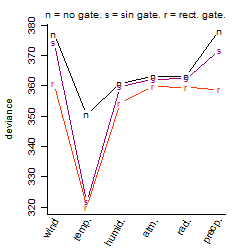
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 317.28 | 0.837 | 5.37 | -1.31 | 3.18 | -2.59 | 1.27 | 2.3 | 0.326 | 15 | 20.8 | 16 | 1.21 | -- |
| 321.95 | 0.836 | 5.4 | -1.64 | 3.21 | -2.58 | 1.26 | -- | 0.239 | 15 | 20.79 | 28 | 1.13 | -- |
| 322.5 | 0.836 | 5.37 | -1.28 | 3.21 | -2.63 | -- | 2.37 | 0.324 | 14.9 | 20.8 | 16 | 1.11 | -- |
| 327.53 | 0.843 | 5.41 | -1.63 | 3.24 | -2.57 | -- | -- | 0.223 | 14.9 | 20.8 | 24 | 1.07 | -- |
| 366.6 | 0.892 | 5.35 | -1.68 | -- | -4.99 | -- | 0.382 | 0.304 | -- | 7.791 | 80 | 2.78 | -- |
| 416.82 | 0.957 | 5.56 | -2.07 | 3.94 | -- | 1.61 | -- | -0.296 | 16.1 | -- | -- | -- | -- |
| 424.24 | 0.965 | 5.55 | -2.05 | 3.95 | -- | -- | -- | -0.287 | 16 | -- | -- | -- | -- |
| 360.51 | 0.884 | 5.43 | -1.67 | -- | -4.75 | -- | -- | 0.105 | -- | 6.156 | 81 | 2.77 | -- |
| 396.02 | 0.927 | 5.35 | -- | 3.27 | -2.89 | -- | -- | 0.441 | 14.8 | 20.8 | 57 | 1.24 | -- |
| 1273.75 | 1.67 | 5.54 | -2.21 | -- | -- | -- | -- | -0.392 | -- | -- | -- | -- | -- |
| 534.95 | 1.08 | 5.5 | -- | 3.99 | -- | -- | -- | -0.0587 | 16 | -- | -- | -- | -- |
| 429.3 | 0.965 | 5.37 | -- | -- | -5.01 | -- | -- | 0.353 | -- | 9.058 | 96 | 2.9 | -- |
| 1402.63 | 1.75 | 5.48 | -- | -- | -- | -- | -- | -0.147 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram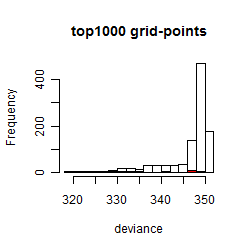 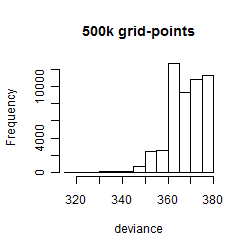
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 319.93 | temperature | 30 | 90 | < th | dose dependent | rect. | 23 | 14 |
| 2 | 320.81 | temperature | 30 | 90 | < th | dose dependent | rect. | 23 | 16 |
| 3 | 321.70 | temperature | 20 | 10 | > th | dose dependent | sin | 7 | NA |
| 9 | 324.79 | temperature | 20 | 270 | > th | dose dependent | rect. | 18 | 16 |
| 19 | 328.45 | temperature | 30 | 10 | < th | dose dependent | sin | 6 | NA |
| 20 | 329.64 | temperature | 30 | 270 | < th | dose dependent | rect. | 21 | 14 |
| 21 | 329.74 | temperature | 30 | 270 | < th | dose dependent | rect. | 21 | 16 |
| 27 | 330.54 | temperature | 30 | 10 | < th | dose dependent | rect. | 23 | 13 |
| 37 | 331.53 | temperature | 30 | 10 | < th | dose dependent | rect. | 23 | 18 |
| 45 | 333.14 | temperature | 25 | 30 | < th | dose independent | rect. | 22 | 14 |
| 61 | 334.66 | temperature | 15 | 270 | > th | dose dependent | rect. | 2 | 9 |
| 135 | 340.58 | temperature | 20 | 10 | > th | dose dependent | rect. | 20 | 16 |
| 229 | 346.82 | temperature | 25 | 14400 | < th | dose dependent | rect. | 4 | 1 |
| 230 | 346.86 | temperature | 25 | 14400 | < th | dose dependent | rect. | 4 | 4 |
| 235 | 346.98 | temperature | 10 | 14400 | > th | dose dependent | rect. | 4 | 4 |
| 238 | 347.03 | temperature | 15 | 14400 | > th | dose dependent | rect. | 6 | 2 |
| 239 | 347.05 | temperature | 25 | 14400 | < th | dose dependent | rect. | 6 | 1 |
| 291 | 347.52 | temperature | 10 | 14400 | > th | dose dependent | rect. | 4 | 1 |
| 417 | 348.52 | temperature | 25 | 14400 | < th | dose dependent | sin | 7 | NA |
| 603 | 349.31 | temperature | 10 | 14400 | > th | dose dependent | sin | 8 | NA |