Os02g0317000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Hypothetical protein.


FiT-DB / Search/ Help/ Sample detail
|
Os02g0317000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
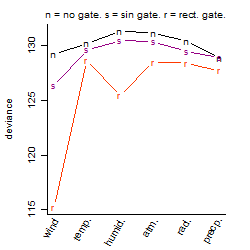
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 128.49 | 0.533 | 2.83 | 0.0575 | 0.119 | 0.244 | -0.142 | -1.48 | 0.26 | 9.05 | 5.718 | 2214 | -- | -- |
| 128.4 | 0.528 | 2.85 | 0.0716 | 0.113 | 0.461 | -0.00438 | -- | 0.263 | 9.26 | 9.499 | 2273 | -- | -- |
| 128.13 | 0.527 | 2.84 | 0.0122 | 0.115 | 0.164 | -- | -1.78 | 0.253 | 8.51 | 10.1 | 2237 | -- | -- |
| 128.27 | 0.527 | 2.86 | 0.071 | 0.109 | 0.521 | -- | -- | 0.253 | 8.53 | 9.824 | 2285 | -- | -- |
| 128.78 | 0.529 | 2.84 | 0.00878 | -- | 0.152 | -- | -1.79 | 0.252 | -- | 10.1 | 2191 | -- | -- |
| 132.33 | 0.539 | 2.87 | -0.0351 | 0.0929 | -- | 0.00352 | -- | 0.203 | 8.2 | -- | -- | -- | -- |
| 132.33 | 0.539 | 2.87 | -0.035 | 0.0929 | -- | -- | -- | 0.203 | 8.21 | -- | -- | -- | -- |
| 128.95 | 0.529 | 2.86 | 0.0709 | -- | 0.508 | -- | -- | 0.252 | -- | 9.801 | 2285 | -- | -- |
| 128.4 | 0.528 | 2.86 | -- | 0.11 | 0.499 | -- | -- | 0.243 | 8.48 | 9.879 | 2285 | -- | -- |
| 132.81 | 0.538 | 2.87 | -0.0343 | -- | -- | -- | -- | 0.204 | -- | -- | -- | -- | -- |
| 132.36 | 0.538 | 2.87 | -- | 0.0927 | -- | -- | -- | 0.207 | 8.23 | -- | -- | -- | -- |
| 129.05 | 0.529 | 2.86 | -- | -- | 0.476 | -- | -- | 0.241 | -- | 10.2 | 2332 | -- | -- |
| 132.84 | 0.538 | 2.87 | -- | -- | -- | -- | -- | 0.208 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 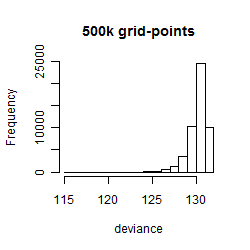
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 115.23 | wind | 7 | 270 | > th | dose dependent | rect. | 8 | 1 |
| 13 | 117.10 | wind | 7 | 270 | > th | dose independent | rect. | 7 | 2 |
| 105 | 124.19 | wind | 1 | 30 | < th | dose dependent | rect. | 9 | 15 |
| 136 | 124.60 | wind | 7 | 10 | > th | dose dependent | rect. | 12 | 1 |
| 179 | 125.03 | wind | 1 | 30 | < th | dose dependent | rect. | 23 | 1 |
| 208 | 125.26 | wind | 9 | 1440 | < th | dose independent | rect. | 1 | 9 |
| 255 | 125.46 | humidity | 90 | 90 | > th | dose independent | rect. | 8 | 2 |
| 256 | 125.46 | wind | 5 | 720 | < th | dose independent | rect. | 1 | 8 |
| 286 | 125.66 | humidity | 90 | 90 | > th | dose dependent | rect. | 8 | 2 |
| 298 | 125.74 | wind | 7 | 1440 | < th | dose independent | rect. | 3 | 6 |
| 336 | 125.90 | wind | 1 | 30 | < th | dose independent | rect. | 9 | 15 |
| 446 | 126.17 | wind | 7 | 1440 | > th | dose independent | rect. | 4 | 1 |
| 463 | 126.20 | wind | 7 | 1440 | > th | dose dependent | rect. | 4 | 1 |
| 488 | 126.26 | wind | 7 | 1440 | < th | dose independent | rect. | 6 | 3 |
| 513 | 126.31 | wind | 1 | 30 | > th | dose independent | sin | 18 | NA |
| 523 | 126.33 | wind | 7 | 4320 | > th | dose dependent | rect. | 6 | 2 |
| 587 | 126.45 | wind | 7 | 1440 | < th | dose independent | rect. | 1 | 4 |
| 630 | 126.51 | wind | 7 | 1440 | < th | dose independent | rect. | 3 | 2 |
| 668 | 126.55 | wind | 1 | 30 | < th | dose dependent | rect. | 13 | 11 |
| 669 | 126.55 | wind | 1 | 30 | < th | dose dependent | rect. | 11 | 13 |
| 670 | 126.56 | wind | 5 | 720 | > th | dose independent | sin | 7 | NA |
| 708 | 126.60 | wind | 5 | 720 | < th | dose independent | sin | 7 | NA |
| 767 | 126.66 | wind | 7 | 720 | < th | dose dependent | rect. | 1 | 10 |
| 890 | 126.84 | wind | 7 | 10 | > th | dose independent | rect. | 12 | 1 |