Os02g0150700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to RING finger protein 12 (LIM domain interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM) (NY-REN-43 antigen).
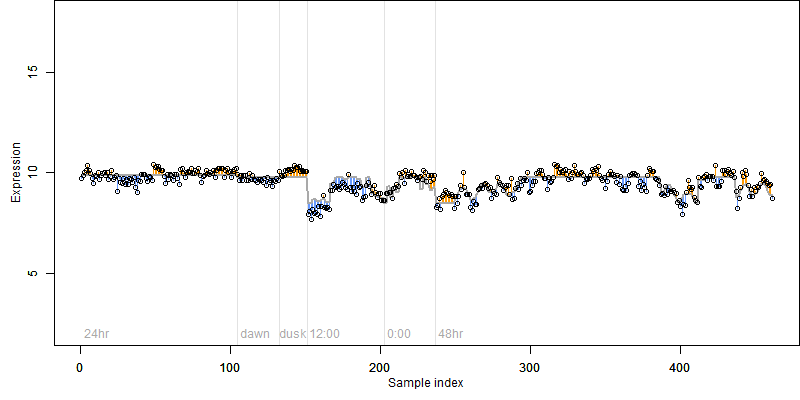

FiT-DB / Search/ Help/ Sample detail
|
Os02g0150700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to RING finger protein 12 (LIM domain interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM) (NY-REN-43 antigen).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
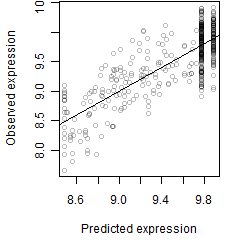
Dependence on each variable

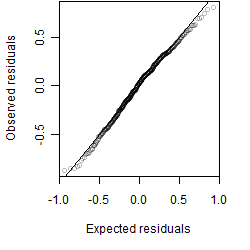
Residual plot
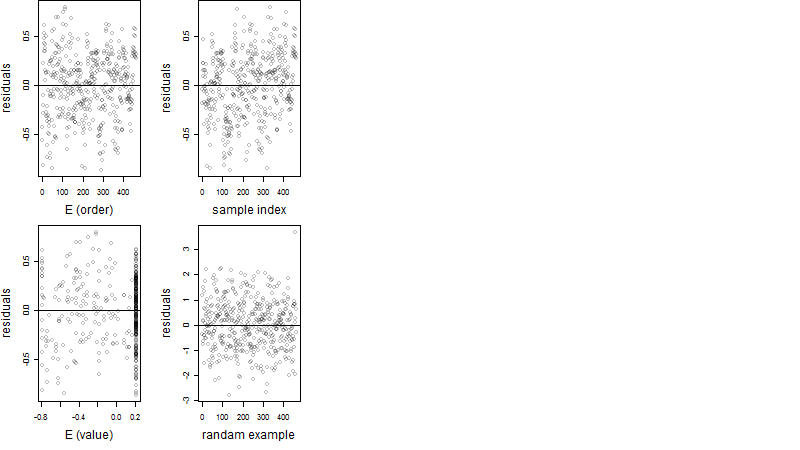
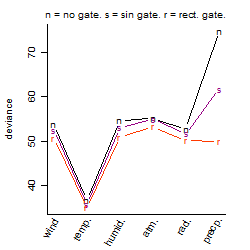
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 35.55 | 0.28 | 9.51 | 0.129 | 0.623 | 1.22 | 0.113 | -0.052 | 0.167 | 23.4 | 21.5 | 1390 | -- | -- |
| 34.6 | 0.274 | 9.51 | 0.212 | 0.525 | 1.14 | 0.239 | -- | 0.168 | 22.9 | 21.81 | 1131 | -- | -- |
| 34.7 | 0.274 | 9.5 | 0.219 | 0.517 | 1.16 | -- | 0.0547 | 0.195 | 23.1 | 21.7 | 1125 | -- | -- |
| 34.33 | 0.273 | 9.5 | 0.216 | 0.505 | 1.13 | -- | -- | 0.179 | 23.5 | 21.9 | 1070 | -- | -- |
| 48.85 | 0.326 | 9.51 | 0.141 | -- | 1.24 | -- | -0.0947 | 0.143 | -- | 21.7 | 943 | -- | -- |
| 80.57 | 0.421 | 9.43 | 0.646 | 0.666 | -- | 0.0176 | -- | 0.527 | 23.6 | -- | -- | -- | -- |
| 80.57 | 0.42 | 9.43 | 0.646 | 0.666 | -- | -- | -- | 0.527 | 23.6 | -- | -- | -- | -- |
| 48.41 | 0.324 | 9.5 | 0.199 | -- | 1.24 | -- | -- | 0.147 | -- | 21.84 | 947 | -- | -- |
| 35.35 | 0.277 | 9.51 | -- | 0.513 | 1.21 | -- | -- | 0.141 | 23.4 | 21.8 | 1117 | -- | -- |
| 108.41 | 0.487 | 9.41 | 0.664 | -- | -- | -- | -- | 0.534 | -- | -- | -- | -- | -- |
| 91.56 | 0.448 | 9.44 | -- | 0.673 | -- | -- | -- | 0.455 | 23.6 | -- | -- | -- | -- |
| 49.34 | 0.327 | 9.51 | -- | -- | 1.29 | -- | -- | 0.111 | -- | 21.89 | 966 | -- | -- |
| 120.04 | 0.511 | 9.43 | -- | -- | -- | -- | -- | 0.461 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram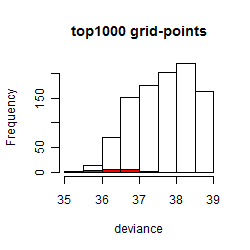 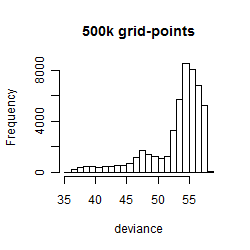
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 35.12 | temperature | 10 | 720 | > th | dose dependent | rect. | 20 | 15 |
| 3 | 35.57 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 1 |
| 5 | 35.73 | temperature | 25 | 720 | < th | dose dependent | rect. | 17 | 20 |
| 10 | 35.83 | temperature | 15 | 720 | > th | dose dependent | sin | 9 | NA |
| 25 | 36.11 | temperature | 25 | 1440 | < th | dose dependent | rect. | 19 | 11 |
| 37 | 36.21 | temperature | 25 | 1440 | < th | dose dependent | rect. | 4 | 1 |
| 43 | 36.30 | temperature | 15 | 1440 | > th | dose dependent | rect. | 4 | 1 |
| 65 | 36.39 | temperature | 25 | 720 | < th | dose dependent | rect. | 24 | 23 |
| 85 | 36.50 | temperature | 25 | 720 | < th | dose dependent | rect. | 21 | 23 |
| 103 | 36.59 | temperature | 15 | 720 | > th | dose dependent | rect. | 20 | 19 |
| 123 | 36.64 | temperature | 25 | 1440 | < th | dose dependent | sin | 11 | NA |
| 142 | 36.71 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 170 | 36.77 | temperature | 15 | 720 | > th | dose dependent | rect. | 15 | 20 |
| 183 | 36.84 | temperature | 15 | 1440 | > th | dose dependent | rect. | 20 | 10 |
| 340 | 37.32 | temperature | 25 | 720 | < th | dose dependent | rect. | 8 | 23 |
| 372 | 37.41 | temperature | 25 | 720 | < th | dose dependent | rect. | 6 | 23 |