Os01g0946300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Lecithin:cholesterol acyltransferase family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os01g0946300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Lecithin:cholesterol acyltransferase family protein.
|
|
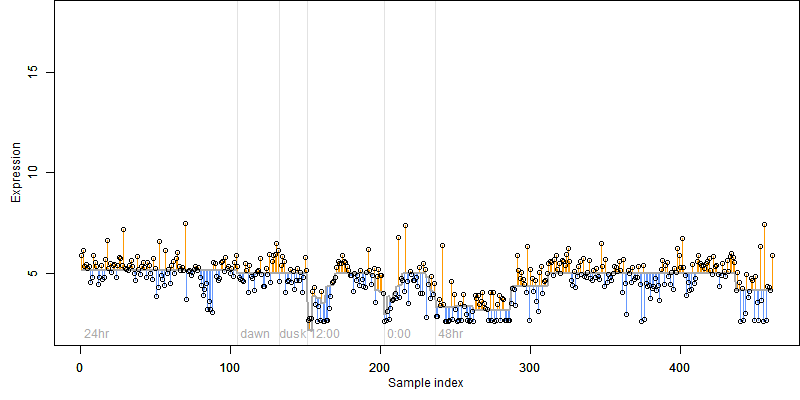
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
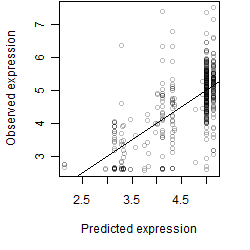
Dependence on each variable

Residual plot

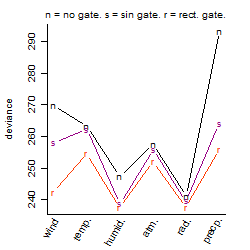
Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 244.31 | 0.734 | 4.59 | 0.548 | 0.0985 | -1.74 | 1.56 | 0.748 | 0.341 | 7.5 | 1009 | 23906 | -- | -- |
| 245.32 | 0.729 | 4.56 | 0.798 | 0.0997 | -1.74 | 1.54 | -- | 0.395 | 7.51 | 1009 | 23818 | -- | -- |
| 249.86 | 0.736 | 4.61 | 0.586 | 0.16 | -1.72 | -- | 0.811 | 0.316 | 10.9 | 1009 | 23895 | -- | -- |
| 250.57 | 0.737 | 4.56 | 0.849 | 0.172 | -1.73 | -- | -- | 0.397 | 11.6 | 1009 | 23782 | -- | -- |
| 251.49 | 0.739 | 4.59 | 0.604 | -- | -1.73 | -- | 0.696 | 0.335 | -- | 1009 | 23911 | -- | -- |
| 339.79 | 0.864 | 4.46 | 1.71 | 0.106 | -- | 1.53 | -- | 0.839 | 7.3 | -- | -- | -- | -- |
| 345.46 | 0.87 | 4.46 | 1.74 | 0.144 | -- | -- | -- | 0.839 | 10.3 | -- | -- | -- | -- |
| 252.44 | 0.74 | 4.56 | 0.84 | -- | -1.73 | -- | -- | 0.398 | -- | 1009 | 23782 | -- | -- |
| 254.11 | 0.742 | 4.61 | -- | 0.175 | -2.53 | -- | -- | 0.143 | 11.5 | 1010 | 32946 | -- | -- |
| 346.73 | 0.87 | 4.46 | 1.74 | -- | -- | -- | -- | 0.838 | -- | -- | -- | -- | -- |
| 425.49 | 0.965 | 4.5 | -- | 0.139 | -- | -- | -- | 0.645 | 9.41 | -- | -- | -- | -- |
| 254.88 | 0.744 | 4.62 | -- | -- | -2.87 | -- | -- | 0.151 | -- | 1013 | 31787 | -- | -- |
| 426.62 | 0.964 | 4.5 | -- | -- | -- | -- | -- | 0.645 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram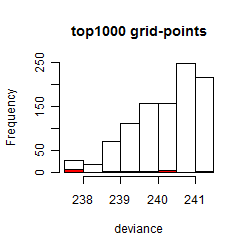 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 237.58 | radiation | 40 | 43200 | < th | dose independent | rect. | 13 | 1 |
| 3 | 237.60 | humidity | 60 | 43200 | < th | dose dependent | rect. | 2 | 12 |
| 6 | 237.61 | humidity | 60 | 43200 | < th | dose dependent | rect. | 4 | 10 |
| 9 | 237.66 | radiation | 40 | 43200 | > th | dose independent | rect. | 12 | 2 |
| 12 | 237.72 | radiation | 30 | 43200 | > th | dose dependent | rect. | 12 | 2 |
| 13 | 237.72 | humidity | 60 | 43200 | < th | dose dependent | rect. | 8 | 6 |
| 53 | 238.75 | humidity | 60 | 43200 | < th | dose dependent | sin | 6 | NA |
| 133 | 239.14 | radiation | 40 | 43200 | < th | dose independent | rect. | 11 | 8 |
| 243 | 239.59 | radiation | 30 | 43200 | > th | dose dependent | sin | 16 | NA |
| 246 | 239.60 | radiation | 40 | 43200 | > th | dose independent | rect. | 11 | 21 |
| 421 | 240.17 | humidity | 70 | 43200 | < th | dose dependent | rect. | 8 | 2 |
| 445 | 240.22 | radiation | 40 | 43200 | > th | dose independent | sin | 18 | NA |
| 462 | 240.26 | radiation | 40 | 43200 | < th | dose independent | sin | 17 | NA |
| 477 | 240.34 | radiation | 30 | 43200 | < th | dose independent | sin | 19 | NA |
| 802 | 241.02 | radiation | 20 | 43200 | > th | dose dependent | no | NA | NA |
| 895 | 241.03 | humidity | 60 | 43200 | > th | dose independent | rect. | 8 | 2 |