Os01g0740400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF1005 family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os01g0740400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF1005 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

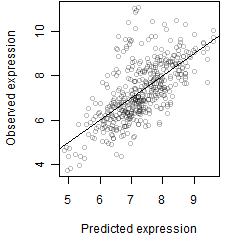 __
__
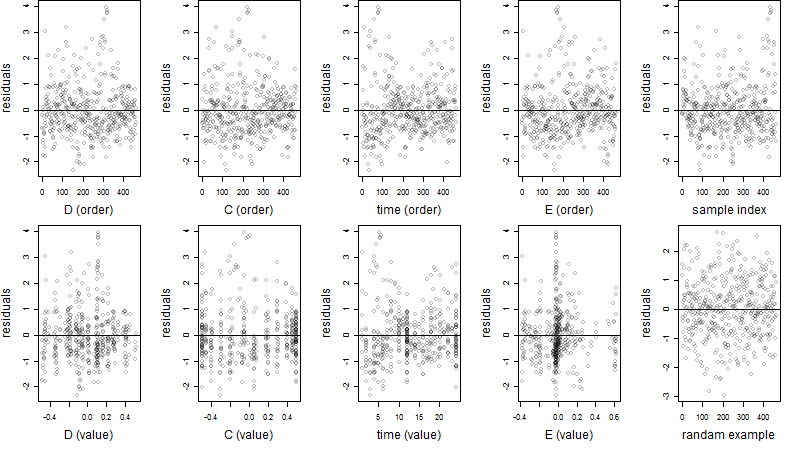
Dependence on each variable

Residual plot
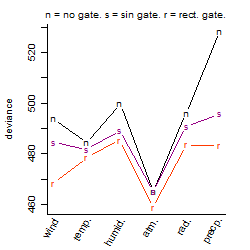
Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 466.31 | 1.01 | 7.44 | -2.27 | 1.72 | -0.177 | 0.0363 | -7.42 | -0.445 | 11.6 | 1008 | 14690 | -- | -- |
| 512.73 | 1.05 | 7.39 | -1.58 | 1.74 | -0.987 | 0.776 | -- | -0.508 | 11.1 | 1001 | 14431 | -- | -- |
| 464.58 | 1 | 7.46 | -2.72 | 1.71 | -0.134 | -- | -7.51 | -0.533 | 11.6 | 1011 | 14089 | -- | -- |
| 503.56 | 1.05 | 7.35 | -1.44 | 1.8 | -1.17 | -- | -- | -0.384 | 11.7 | 999.8 | 11884 | -- | -- |
| 648.69 | 1.19 | 7.51 | -2.87 | -- | -0.0201 | -- | -8.09 | -0.568 | -- | 1011 | 13998 | -- | -- |
| 545.02 | 1.09 | 7.32 | -1.7 | 1.75 | -- | 0.0192 | -- | -0.305 | 11.6 | -- | -- | -- | -- |
| 545.02 | 1.09 | 7.32 | -1.7 | 1.75 | -- | -- | -- | -0.305 | 11.6 | -- | -- | -- | -- |
| 697.5 | 1.23 | 7.36 | -1.53 | -- | -1.24 | -- | -- | -0.42 | -- | 1000 | 11192 | -- | -- |
| 555.46 | 1.1 | 7.37 | -- | 1.76 | -1.56 | -- | -- | -0.327 | 11.8 | 1001 | 11994 | -- | -- |
| 736.83 | 1.27 | 7.36 | -1.75 | -- | -- | -- | -- | -0.325 | -- | -- | -- | -- | -- |
| 621.15 | 1.17 | 7.27 | -- | 1.77 | -- | -- | -- | -0.115 | 11.7 | -- | -- | -- | -- |
| 754.18 | 1.28 | 7.35 | -- | -- | -1.59 | -- | -- | -0.31 | -- | 1000 | 11283 | -- | -- |
| 817.45 | 1.33 | 7.31 | -- | -- | -- | -- | -- | -0.131 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 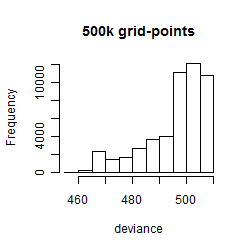
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 458.94 | atmosphere | 1005 | 14400 | < th | dose independent | rect. | 7 | 2 |
| 2 | 459.21 | atmosphere | 1005 | 14400 | > th | dose independent | rect. | 8 | 1 |
| 9 | 460.51 | atmosphere | 1005 | 14400 | > th | dose independent | rect. | 6 | 4 |
| 29 | 463.03 | atmosphere | 995 | 14400 | > th | dose dependent | rect. | 23 | 1 |
| 33 | 463.18 | atmosphere | 1015 | 14400 | < th | dose dependent | rect. | 23 | 1 |
| 88 | 464.78 | atmosphere | 1010 | 14400 | < th | dose dependent | rect. | 8 | 8 |
| 90 | 464.79 | atmosphere | 1010 | 14400 | < th | dose dependent | rect. | 8 | 6 |
| 95 | 464.82 | atmosphere | 1010 | 14400 | < th | dose dependent | rect. | 7 | 17 |
| 131 | 464.95 | atmosphere | 1010 | 14400 | < th | dose dependent | rect. | 20 | 20 |
| 147 | 464.99 | atmosphere | 1010 | 14400 | < th | dose dependent | rect. | 11 | 1 |
| 194 | 465.08 | atmosphere | 1010 | 14400 | < th | dose dependent | sin | 23 | NA |
| 237 | 465.12 | atmosphere | 1010 | 14400 | < th | dose dependent | rect. | 13 | 23 |
| 238 | 465.12 | atmosphere | 1010 | 14400 | < th | dose dependent | rect. | 15 | 23 |
| 240 | 465.13 | atmosphere | 1010 | 14400 | < th | dose dependent | no | NA | NA |
| 263 | 465.15 | atmosphere | 1010 | 14400 | < th | dose dependent | rect. | 15 | 21 |
| 287 | 465.18 | atmosphere | 1010 | 14400 | < th | dose dependent | rect. | 11 | 23 |
| 292 | 465.19 | atmosphere | 995 | 14400 | > th | dose dependent | rect. | 9 | 15 |
| 460 | 465.39 | atmosphere | 1010 | 14400 | < th | dose dependent | sin | 10 | NA |
| 485 | 465.42 | atmosphere | 995 | 14400 | > th | dose dependent | rect. | 11 | 1 |
| 494 | 465.43 | atmosphere | 995 | 14400 | > th | dose dependent | sin | 14 | NA |
| 509 | 465.44 | atmosphere | 995 | 14400 | > th | dose dependent | rect. | 13 | 11 |
| 559 | 465.48 | atmosphere | 995 | 14400 | > th | dose dependent | rect. | 19 | 20 |
| 577 | 465.50 | atmosphere | 995 | 14400 | > th | dose dependent | rect. | 10 | 4 |
| 737 | 465.60 | atmosphere | 1010 | 14400 | < th | dose dependent | rect. | 13 | 1 |
| 787 | 465.63 | atmosphere | 995 | 14400 | > th | dose dependent | rect. | 13 | 1 |
| 820 | 465.64 | atmosphere | 995 | 14400 | > th | dose dependent | rect. | 13 | 23 |
| 823 | 465.65 | atmosphere | 995 | 14400 | > th | dose dependent | no | NA | NA |