Os01g0732000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Mitochondrial import receptor subunit TOM7-1 (Translocase of outer membrane 7 kDa subunit 1).


FiT-DB / Search/ Help/ Sample detail
|
Os01g0732000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Mitochondrial import receptor subunit TOM7-1 (Translocase of outer membrane 7 kDa subunit 1).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
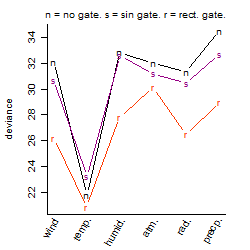
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21.72 | 0.219 | 11.3 | -0.441 | 0.433 | 1.15 | 0.236 | 0.635 | -0.298 | 11.8 | 19.86 | 679 | -- | -- |
| 21.95 | 0.218 | 11.3 | -0.566 | 0.398 | 1.2 | 0.147 | -- | -0.334 | 11.2 | 20.44 | 605 | -- | -- |
| 21.93 | 0.218 | 11.3 | -0.451 | 0.419 | 1.18 | -- | 0.541 | -0.301 | 11.7 | 19.66 | 649 | -- | -- |
| 22.04 | 0.219 | 11.3 | -0.556 | 0.395 | 1.2 | -- | -- | -0.334 | 11.2 | 20.4 | 602 | -- | -- |
| 24.92 | 0.232 | 11.3 | -0.464 | -- | 1.17 | -- | 0.299 | -0.304 | -- | 19.68 | 234 | -- | -- |
| 49.17 | 0.329 | 11.3 | -0.256 | 0.402 | -- | 0.163 | -- | -0.119 | 15.5 | -- | -- | -- | -- |
| 49.25 | 0.329 | 11.3 | -0.253 | 0.403 | -- | -- | -- | -0.118 | 15.5 | -- | -- | -- | -- |
| 24.95 | 0.233 | 11.3 | -0.519 | -- | 1.17 | -- | -- | -0.317 | -- | 20.02 | 233 | -- | -- |
| 29.25 | 0.252 | 11.3 | -- | 0.332 | 1.02 | -- | -- | -0.241 | 11.3 | 21.4 | 530 | -- | -- |
| 58.33 | 0.357 | 11.3 | -0.27 | -- | -- | -- | -- | -0.129 | -- | -- | -- | -- | -- |
| 50.94 | 0.334 | 11.3 | -- | 0.408 | -- | -- | -- | -0.0901 | 15.5 | -- | -- | -- | -- |
| 31.56 | 0.262 | 11.3 | -- | -- | 1.03 | -- | -- | -0.24 | -- | 20.85 | 229 | -- | -- |
| 60.25 | 0.362 | 11.3 | -- | -- | -- | -- | -- | -0.099 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 20.85 | temperature | 10 | 720 | > th | dose dependent | rect. | 3 | 21 |
| 2 | 20.98 | temperature | 10 | 720 | > th | dose dependent | rect. | 12 | 23 |
| 6 | 21.17 | temperature | 20 | 720 | > th | dose dependent | rect. | 19 | 22 |
| 8 | 21.22 | temperature | 10 | 720 | > th | dose dependent | rect. | 12 | 21 |
| 19 | 21.35 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 23 |
| 58 | 21.74 | temperature | 20 | 720 | > th | dose dependent | rect. | 23 | 23 |
| 62 | 21.76 | temperature | 20 | 720 | > th | dose dependent | no | NA | NA |
| 122 | 22.14 | temperature | 20 | 720 | > th | dose dependent | rect. | 8 | 23 |
| 250 | 22.96 | temperature | 30 | 720 | < th | dose dependent | rect. | 24 | 21 |
| 283 | 23.14 | temperature | 20 | 1440 | > th | dose dependent | rect. | 11 | 15 |
| 301 | 23.21 | temperature | 20 | 1440 | > th | dose dependent | sin | 17 | NA |
| 406 | 23.40 | temperature | 25 | 1440 | > th | dose dependent | rect. | 12 | 1 |
| 478 | 23.50 | temperature | 20 | 1440 | > th | dose dependent | rect. | 15 | 7 |
| 483 | 23.51 | temperature | 30 | 1440 | < th | dose dependent | rect. | 18 | 1 |
| 501 | 23.55 | temperature | 20 | 1440 | > th | dose dependent | rect. | 17 | 4 |
| 549 | 23.63 | temperature | 30 | 720 | < th | dose dependent | rect. | 6 | 23 |
| 626 | 23.76 | temperature | 20 | 1440 | > th | dose dependent | rect. | 18 | 1 |
| 678 | 23.81 | temperature | 30 | 720 | < th | dose dependent | rect. | 13 | 23 |
| 686 | 23.83 | temperature | 25 | 1440 | > th | dose dependent | rect. | 11 | 4 |
| 696 | 23.84 | temperature | 30 | 720 | < th | dose dependent | no | NA | NA |