Os01g0722700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Hexokinase.


FiT-DB / Search/ Help/ Sample detail
|
Os01g0722700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Hexokinase.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

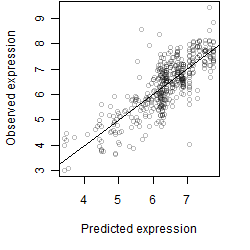
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 177.42 | 0.626 | 6.2 | -0.251 | 0.689 | -3.47 | -0.933 | -0.915 | 0.819 | 19.7 | 23.43 | 988 | -- | -- |
| 177.26 | 0.62 | 6.21 | -0.394 | 0.725 | -3.42 | -0.83 | -- | 0.826 | 19.8 | 23.08 | 1056 | -- | -- |
| 177.97 | 0.621 | 6.2 | -0.261 | 0.746 | -3.67 | -- | -1.04 | 0.834 | 19.8 | 23 | 1076 | -- | -- |
| 179.29 | 0.624 | 6.22 | -0.392 | 0.737 | -3.46 | -- | -- | 0.821 | 19.8 | 23 | 1048 | -- | -- |
| 200.95 | 0.66 | 6.19 | -0.306 | -- | -3.67 | -- | -0.679 | 0.808 | -- | 23.07 | 834 | -- | -- |
| 321.41 | 0.84 | 6.1 | 0.547 | 0.899 | -- | -0.825 | -- | 1.38 | 21 | -- | -- | -- | -- |
| 323.76 | 0.843 | 6.1 | 0.575 | 0.901 | -- | -- | -- | 1.38 | 21 | -- | -- | -- | -- |
| 201.87 | 0.662 | 6.2 | -0.381 | -- | -3.56 | -- | -- | 0.794 | -- | 23.1 | 821 | -- | -- |
| 182.52 | 0.629 | 6.2 | -- | 0.725 | -3.24 | -- | -- | 0.88 | 19.8 | 23.1 | 1016 | -- | -- |
| 370.44 | 0.899 | 6.07 | 0.576 | -- | -- | -- | -- | 1.37 | -- | -- | -- | -- | -- |
| 332.48 | 0.853 | 6.11 | -- | 0.901 | -- | -- | -- | 1.31 | 21.1 | -- | -- | -- | -- |
| 205.19 | 0.667 | 6.19 | -- | -- | -3.37 | -- | -- | 0.857 | -- | 23.12 | 802 | -- | -- |
| 379.21 | 0.909 | 6.09 | -- | -- | -- | -- | -- | 1.31 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance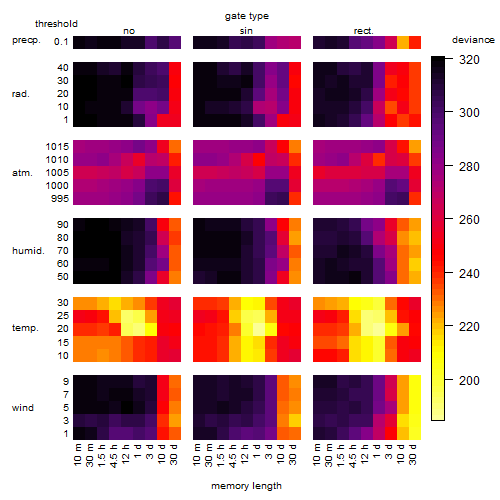
|
Histogram 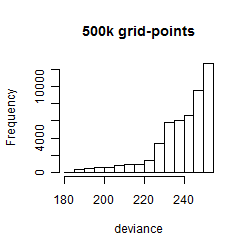
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 184.40 | temperature | 20 | 1440 | > th | dose independent | rect. | 21 | 17 |
| 5 | 184.80 | temperature | 20 | 1440 | < th | dose independent | rect. | 21 | 17 |
| 9 | 185.05 | temperature | 20 | 1440 | > th | dose independent | sin | 5 | NA |
| 16 | 185.28 | temperature | 20 | 1440 | < th | dose independent | sin | 5 | NA |
| 41 | 186.14 | temperature | 20 | 1440 | > th | dose independent | rect. | 1 | 12 |
| 43 | 186.18 | temperature | 20 | 1440 | < th | dose independent | rect. | 1 | 12 |
| 69 | 186.66 | temperature | 20 | 1440 | < th | dose independent | rect. | 3 | 8 |
| 80 | 186.87 | temperature | 20 | 1440 | > th | dose independent | rect. | 3 | 8 |
| 200 | 188.91 | temperature | 25 | 720 | < th | dose dependent | rect. | 1 | 20 |
| 204 | 188.94 | temperature | 25 | 720 | < th | dose dependent | rect. | 23 | 21 |
| 232 | 189.31 | temperature | 20 | 1440 | > th | dose independent | rect. | 1 | 23 |
| 234 | 189.32 | temperature | 20 | 1440 | > th | dose independent | rect. | 11 | 23 |
| 238 | 189.37 | temperature | 20 | 1440 | > th | dose independent | no | NA | NA |
| 244 | 189.42 | temperature | 20 | 1440 | > th | dose independent | rect. | 8 | 23 |
| 264 | 189.70 | temperature | 20 | 1440 | > th | dose independent | rect. | 3 | 23 |
| 268 | 189.71 | temperature | 20 | 1440 | < th | dose independent | rect. | 11 | 23 |
| 272 | 189.75 | temperature | 20 | 1440 | < th | dose independent | rect. | 1 | 23 |
| 277 | 189.80 | temperature | 20 | 1440 | < th | dose independent | no | NA | NA |
| 282 | 189.83 | temperature | 20 | 1440 | < th | dose independent | rect. | 8 | 23 |
| 320 | 190.28 | temperature | 20 | 1440 | < th | dose dependent | rect. | 4 | 1 |
| 429 | 191.69 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 16 |
| 512 | 192.52 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 513 | 192.52 | temperature | 25 | 720 | < th | dose dependent | rect. | 13 | 23 |
| 575 | 193.28 | temperature | 25 | 720 | < th | dose dependent | rect. | 4 | 23 |
| 593 | 193.49 | temperature | 20 | 1440 | < th | dose independent | rect. | 5 | 3 |
| 613 | 193.69 | temperature | 25 | 1440 | < th | dose dependent | sin | 8 | NA |
| 693 | 194.39 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 2 |
| 720 | 194.65 | temperature | 25 | 1440 | < th | dose dependent | rect. | 18 | 12 |
| 742 | 194.77 | temperature | 25 | 720 | < th | dose dependent | rect. | 6 | 23 |
| 748 | 194.83 | temperature | 25 | 720 | < th | dose dependent | rect. | 8 | 23 |
| 858 | 195.56 | temperature | 25 | 720 | < th | dose dependent | sin | 2 | NA |