Os01g0634300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Heat shock protein DnaJ, N-terminal domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os01g0634300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Heat shock protein DnaJ, N-terminal domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

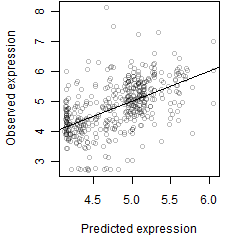
Dependence on each variable

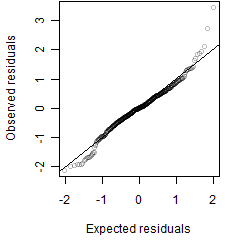
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 214.89 | 0.689 | 4.57 | 1.65 | 0.517 | -2.49 | 0.0312 | 2.84 | 0.916 | 23.2 | 14.64 | 963 | -- | -- |
| 222.09 | 0.694 | 4.65 | 1.18 | 0.402 | -2.26 | 0.381 | -- | 0.8 | 0.563 | 14.9 | 1113 | -- | -- |
| 214.87 | 0.683 | 4.57 | 1.66 | 0.51 | -2.47 | -- | 2.89 | 0.922 | 23.2 | 14.5 | 979 | -- | -- |
| 221.85 | 0.694 | 4.65 | 1.08 | 0.573 | -2.53 | -- | -- | 0.767 | 22.4 | 13.58 | 874 | -- | -- |
| 222.55 | 0.695 | 4.55 | 1.72 | -- | -2.19 | -- | 3 | 0.929 | -- | 13.44 | 1311 | -- | -- |
| 308.16 | 0.823 | 4.72 | 0.408 | 0.0852 | -- | 0.43 | -- | 0.366 | 1.37 | -- | -- | -- | -- |
| 308.88 | 0.823 | 4.72 | 0.394 | 0.0867 | -- | -- | -- | 0.364 | 0.329 | -- | -- | -- | -- |
| 229.72 | 0.706 | 4.63 | 1.08 | -- | -2.15 | -- | -- | 0.763 | -- | 13.7 | 1429 | -- | -- |
| 248.54 | 0.734 | 4.69 | -- | 0.49 | -2.05 | -- | -- | 0.583 | 22.4 | 12.91 | 859 | -- | -- |
| 309.36 | 0.822 | 4.72 | 0.397 | -- | -- | -- | -- | 0.365 | -- | -- | -- | -- | -- |
| 312.98 | 0.828 | 4.73 | -- | 0.0925 | -- | -- | -- | 0.32 | 0.552 | -- | -- | -- | -- |
| 256.34 | 0.746 | 4.68 | -- | -- | -1.71 | -- | -- | 0.573 | -- | 13.93 | 1289 | -- | -- |
| 313.51 | 0.826 | 4.73 | -- | -- | -- | -- | -- | 0.321 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 213.84 | temperature | 10 | 720 | > th | dose dependent | rect. | 13 | 23 |
| 2 | 214.05 | temperature | 10 | 720 | > th | dose dependent | rect. | 24 | 22 |
| 5 | 214.30 | temperature | 30 | 1440 | < th | dose dependent | rect. | 13 | 6 |
| 8 | 214.48 | temperature | 30 | 1440 | < th | dose dependent | rect. | 14 | 4 |
| 9 | 214.56 | temperature | 15 | 1440 | > th | dose dependent | rect. | 15 | 1 |
| 13 | 214.62 | temperature | 30 | 1440 | < th | dose dependent | rect. | 11 | 8 |
| 14 | 214.63 | temperature | 30 | 1440 | < th | dose dependent | rect. | 9 | 10 |
| 38 | 215.22 | temperature | 10 | 1440 | > th | dose dependent | rect. | 11 | 5 |
| 42 | 215.38 | temperature | 10 | 720 | > th | dose dependent | rect. | 8 | 23 |
| 48 | 215.52 | temperature | 30 | 1440 | < th | dose dependent | sin | 21 | NA |
| 57 | 215.70 | temperature | 30 | 1440 | < th | dose dependent | rect. | 15 | 1 |
| 80 | 216.00 | temperature | 10 | 720 | > th | dose dependent | rect. | 4 | 23 |
| 167 | 216.77 | temperature | 15 | 720 | > th | dose dependent | no | NA | NA |
| 181 | 216.84 | temperature | 10 | 1440 | > th | dose dependent | sin | 21 | NA |
| 190 | 216.89 | temperature | 30 | 720 | < th | dose dependent | rect. | 2 | 22 |
| 237 | 217.16 | temperature | 10 | 1440 | > th | dose dependent | rect. | 11 | 14 |
| 320 | 217.57 | temperature | 15 | 720 | > th | dose dependent | rect. | 18 | 23 |
| 343 | 217.68 | temperature | 30 | 720 | < th | dose dependent | rect. | 11 | 23 |
| 386 | 217.89 | temperature | 15 | 720 | > th | dose dependent | rect. | 2 | 23 |
| 395 | 217.93 | temperature | 30 | 720 | < th | dose dependent | rect. | 20 | 23 |
| 538 | 218.68 | temperature | 30 | 720 | < th | dose dependent | no | NA | NA |
| 583 | 218.89 | temperature | 30 | 720 | < th | dose dependent | rect. | 22 | 23 |
| 654 | 219.15 | temperature | 30 | 720 | < th | dose dependent | rect. | 6 | 23 |