Os01g0558200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Glu/Leu/Phe/Val dehydrogenase family protein.
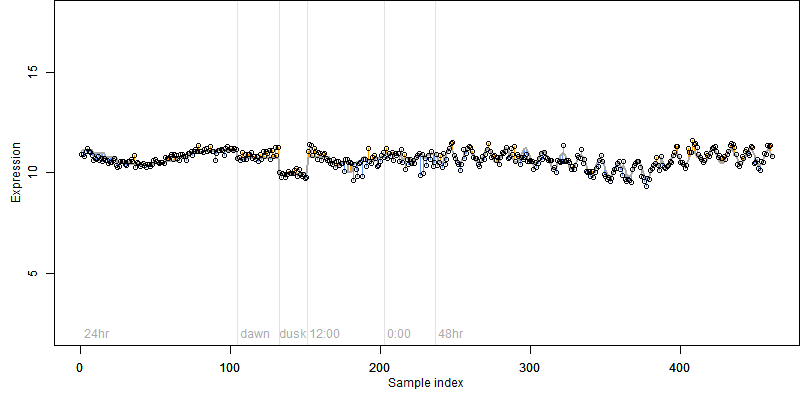

FiT-DB / Search/ Help/ Sample detail
|
Os01g0558200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Glu/Leu/Phe/Val dehydrogenase family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable
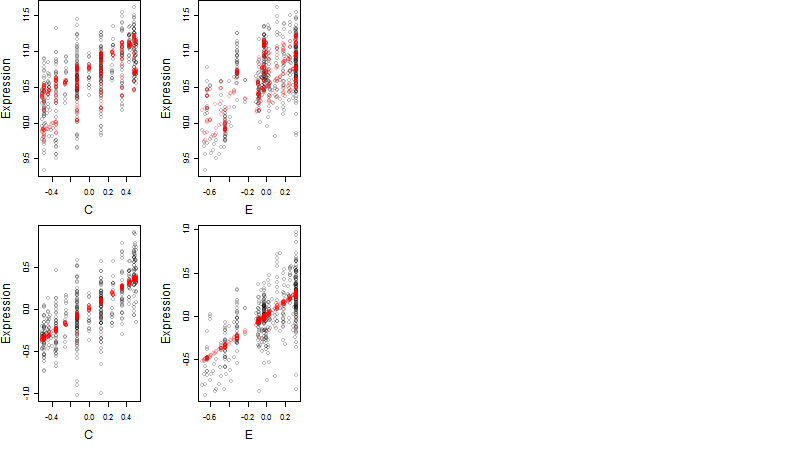
Residual plot

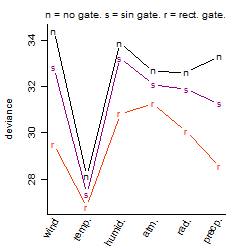
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 26.87 | 0.244 | 10.6 | -0.0667 | 0.722 | 0.773 | 0.466 | 0.129 | 0.171 | 4.98 | 28.6 | 4578 | -- | -- |
| 26.81 | 0.241 | 10.6 | -0.0237 | 0.718 | 0.786 | 0.463 | -- | 0.188 | 4.99 | 28.39 | 4595 | -- | -- |
| 27.36 | 0.244 | 10.6 | -0.0674 | 0.712 | 0.772 | -- | 0.19 | 0.175 | 4.96 | 28.4 | 4622 | -- | -- |
| 27.35 | 0.244 | 10.6 | -0.0252 | 0.721 | 0.782 | -- | -- | 0.187 | 5 | 28.35 | 4600 | -- | -- |
| 52.65 | 0.338 | 10.6 | -0.0551 | -- | 0.838 | -- | 0.271 | 0.197 | -- | 28.2 | 4833 | -- | -- |
| 48.21 | 0.325 | 10.6 | -0.278 | 0.733 | -- | 0.422 | -- | 0.122 | 5.12 | -- | -- | -- | -- |
| 48.66 | 0.327 | 10.6 | -0.278 | 0.736 | -- | -- | -- | 0.12 | 5.12 | -- | -- | -- | -- |
| 52.49 | 0.337 | 10.6 | 0.0228 | -- | 0.868 | -- | -- | 0.223 | -- | 28.29 | 4841 | -- | -- |
| 27.37 | 0.244 | 10.6 | -- | 0.721 | 0.787 | -- | -- | 0.19 | 4.99 | 28.33 | 4600 | -- | -- |
| 77.05 | 0.41 | 10.6 | -0.252 | -- | -- | -- | -- | 0.139 | -- | -- | -- | -- | -- |
| 50.7 | 0.333 | 10.6 | -- | 0.731 | -- | -- | -- | 0.151 | 5.13 | -- | -- | -- | -- |
| 52.48 | 0.337 | 10.6 | -- | -- | 0.858 | -- | -- | 0.215 | -- | 28.36 | 4841 | -- | -- |
| 78.73 | 0.414 | 10.6 | -- | -- | -- | -- | -- | 0.167 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 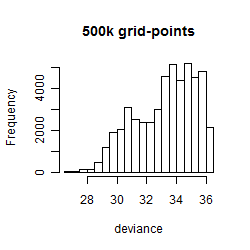
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 26.78 | temperature | 25.0 | 4320 | > th | dose dependent | rect. | 7 | 3 |
| 51 | 27.33 | temperature | 25.0 | 4320 | > th | dose dependent | sin | 5 | NA |
| 65 | 27.48 | temperature | 25.0 | 4320 | > th | dose independent | rect. | 7 | 1 |
| 210 | 28.07 | temperature | 25.0 | 4320 | < th | dose independent | rect. | 7 | 1 |
| 254 | 28.11 | temperature | 25.0 | 4320 | > th | dose dependent | no | NA | NA |
| 344 | 28.54 | precipitation | 0.1 | 43200 | > th | dose dependent | rect. | 17 | 4 |
| 424 | 28.66 | temperature | 25.0 | 14400 | > th | dose dependent | rect. | 18 | 1 |
| 425 | 28.66 | temperature | 25.0 | 14400 | > th | dose dependent | rect. | 17 | 3 |
| 447 | 28.68 | temperature | 30.0 | 14400 | > th | dose independent | rect. | 15 | 1 |
| 448 | 28.68 | temperature | 30.0 | 14400 | < th | dose independent | rect. | 15 | 1 |
| 477 | 28.70 | temperature | 30.0 | 14400 | < th | dose independent | rect. | 14 | 20 |
| 531 | 28.74 | temperature | 30.0 | 14400 | > th | dose independent | rect. | 14 | 20 |
| 546 | 28.76 | temperature | 25.0 | 14400 | < th | dose independent | rect. | 19 | 1 |
| 596 | 28.80 | temperature | 10.0 | 4320 | > th | dose dependent | rect. | 7 | 2 |
| 607 | 28.81 | temperature | 30.0 | 4320 | < th | dose dependent | rect. | 7 | 2 |
| 608 | 28.81 | temperature | 10.0 | 4320 | > th | dose dependent | rect. | 6 | 4 |
| 671 | 28.89 | temperature | 25.0 | 14400 | < th | dose independent | rect. | 19 | 3 |
| 686 | 28.91 | temperature | 25.0 | 14400 | > th | dose independent | rect. | 19 | 3 |
| 687 | 28.91 | temperature | 25.0 | 14400 | < th | dose independent | rect. | 17 | 6 |
| 690 | 28.91 | temperature | 30.0 | 4320 | < th | dose dependent | rect. | 6 | 4 |
| 706 | 28.93 | temperature | 25.0 | 14400 | > th | dose independent | rect. | 19 | 1 |
| 715 | 28.93 | temperature | 10.0 | 4320 | > th | dose dependent | rect. | 6 | 16 |
| 717 | 28.94 | temperature | 25.0 | 14400 | > th | dose independent | rect. | 17 | 6 |
| 785 | 28.99 | temperature | 30.0 | 4320 | < th | dose dependent | rect. | 18 | 1 |
| 848 | 29.04 | temperature | 30.0 | 14400 | < th | dose independent | rect. | 14 | 5 |
| 894 | 29.06 | temperature | 10.0 | 4320 | > th | dose dependent | rect. | 18 | 1 |
| 916 | 29.08 | temperature | 30.0 | 14400 | > th | dose independent | rect. | 14 | 5 |
| 984 | 29.11 | temperature | 20.0 | 14400 | > th | dose dependent | rect. | 21 | 1 |
| 997 | 29.12 | temperature | 10.0 | 4320 | > th | dose dependent | sin | 20 | NA |