Os01g0343100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF594 family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os01g0343100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF594 family protein.
|
|
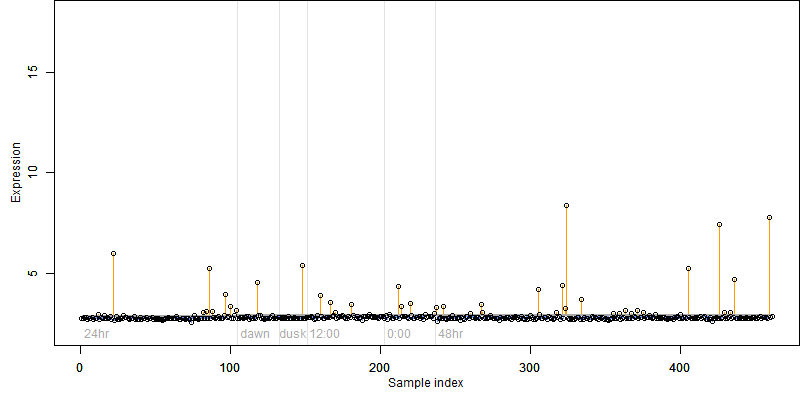
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
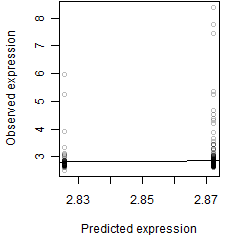
Dependence on each variable

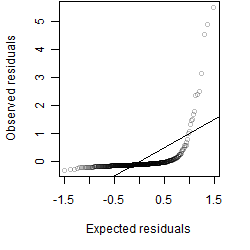
Residual plot

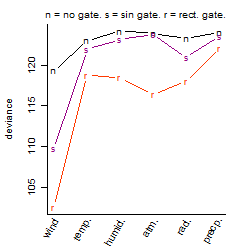
Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 116.59 | 0.507 | 2.83 | 0.0776 | 0.0529 | 0.0897 | 0.362 | 3.75 | -0.0147 | 13.7 | 0.7008 | 98 | -- | -- |
| 121.64 | 0.514 | 2.85 | -0.0314 | 0.0398 | 0.632 | 0.186 | -- | 0.013 | 14.3 | 0.7005 | 98 | -- | -- |
| 116.79 | 0.503 | 2.83 | 0.0961 | 0.088 | 0.0158 | -- | 3.71 | -0.0189 | 9.52 | 0.7309 | 98 | -- | -- |
| 121.62 | 0.514 | 2.86 | -0.00648 | 0.0704 | 0.625 | -- | -- | -0.00605 | 12.5 | 0.7284 | 98 | -- | -- |
| 117.23 | 0.504 | 2.84 | 0.0894 | -- | 0.0599 | -- | 3.6 | -0.0164 | -- | 0.7257 | 98 | -- | -- |
| 125.27 | 0.525 | 2.87 | 0.0667 | 0.115 | -- | 0.142 | -- | -0.04 | 7.7 | -- | -- | -- | -- |
| 125.33 | 0.524 | 2.87 | 0.0701 | 0.116 | -- | -- | -- | -0.0403 | 7.53 | -- | -- | -- | -- |
| 121.9 | 0.514 | 2.86 | -0.0019 | -- | 0.611 | -- | -- | -0.00131 | -- | 0.7291 | 98 | -- | -- |
| 121.61 | 0.514 | 2.86 | -- | 0.0678 | 0.624 | -- | -- | 0.000581 | 12 | 0.746 | 98 | -- | -- |
| 126.05 | 0.525 | 2.87 | 0.0718 | -- | -- | -- | -- | -0.0385 | -- | -- | -- | -- | -- |
| 125.46 | 0.524 | 2.87 | -- | 0.116 | -- | -- | -- | -0.0481 | 7.49 | -- | -- | -- | -- |
| 121.9 | 0.514 | 2.86 | -- | -- | 0.611 | -- | -- | -0.0013 | -- | 0.7289 | 98 | -- | -- |
| 126.18 | 0.524 | 2.87 | -- | -- | -- | -- | -- | -0.0465 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram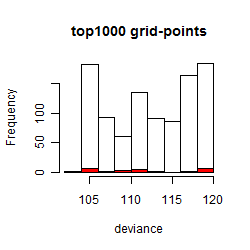 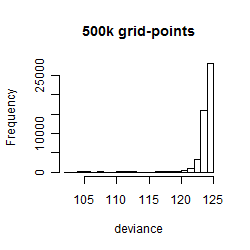
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 102.65 | wind | 1 | 90 | < th | dose dependent | rect. | 4 | 1 |
| 2 | 104.30 | wind | 1 | 90 | < th | dose independent | rect. | 12 | 1 |
| 3 | 104.30 | wind | 1 | 10 | < th | dose independent | rect. | 13 | 1 |
| 11 | 104.30 | wind | 1 | 10 | < th | dose dependent | rect. | 13 | 1 |
| 19 | 104.32 | wind | 1 | 90 | < th | dose dependent | rect. | 12 | 1 |
| 104 | 104.92 | wind | 1 | 90 | < th | dose independent | rect. | 4 | 1 |
| 105 | 104.93 | wind | 1 | 90 | < th | dose independent | rect. | 8 | 7 |
| 187 | 106.29 | wind | 1 | 30 | < th | dose independent | rect. | 10 | 5 |
| 259 | 107.79 | wind | 1 | 10 | < th | dose dependent | rect. | 6 | 9 |
| 299 | 108.52 | wind | 1 | 90 | < th | dose dependent | rect. | 4 | 11 |
| 331 | 109.77 | wind | 1 | 90 | < th | dose independent | sin | 22 | NA |
| 334 | 109.94 | wind | 1 | 30 | < th | dose dependent | sin | 22 | NA |
| 353 | 110.18 | wind | 1 | 10 | < th | dose independent | sin | 21 | NA |
| 366 | 110.42 | wind | 1 | 90 | < th | dose dependent | sin | 0 | NA |
| 455 | 111.56 | wind | 1 | 90 | > th | dose independent | sin | 22 | NA |
| 463 | 111.77 | wind | 1 | 10 | > th | dose independent | sin | 21 | NA |
| 471 | 111.94 | wind | 1 | 90 | < th | dose independent | rect. | 4 | 11 |
| 517 | 112.58 | wind | 1 | 10 | < th | dose dependent | rect. | 6 | 17 |
| 675 | 116.40 | atmosphere | 1005 | 90 | > th | dose dependent | rect. | 5 | 1 |
| 752 | 117.29 | wind | 1 | 90 | < th | dose dependent | rect. | 12 | 17 |
| 816 | 118.01 | radiation | 10 | 30 | < th | dose independent | rect. | 9 | 1 |
| 821 | 118.07 | wind | 1 | 90 | > th | dose independent | rect. | 7 | 12 |
| 864 | 118.32 | wind | 1 | 10 | > th | dose independent | rect. | 7 | 12 |
| 887 | 118.45 | humidity | 90 | 10 | > th | dose dependent | rect. | 6 | 1 |
| 941 | 118.71 | temperature | 20 | 90 | < th | dose independent | rect. | 5 | 1 |
| 944 | 118.72 | temperature | 20 | 10 | < th | dose independent | rect. | 5 | 1 |
| 999 | 118.89 | temperature | 20 | 90 | < th | dose independent | rect. | 4 | 18 |