Os01g0308600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.
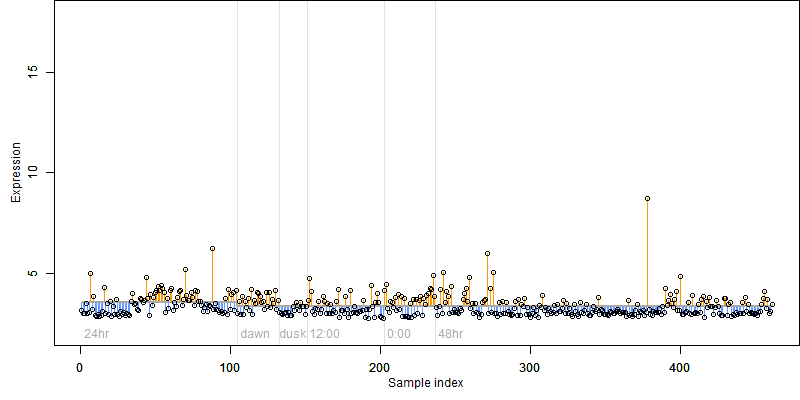

FiT-DB / Search/ Help/ Sample detail
|
Os01g0308600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
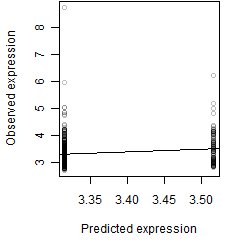
Dependence on each variable

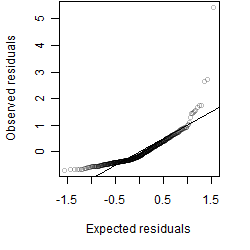
Residual plot

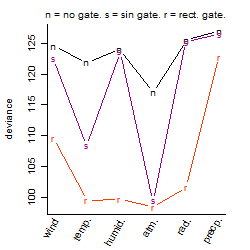
Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 112.61 | 0.494 | -150 | 936 | 0.338 | -38400 | 0.0657 | 235000 | 0.189 | 0.619 | 995 | 90 | -- | -- |
| 126.89 | 0.529 | 3.33 | -0.191 | 0.332 | 1.48 | 0.0725 | -- | 0.182 | 0.175 | 995 | 90 | -- | -- |
| 112.62 | 0.494 | -150 | 936 | 0.338 | -38400 | -- | 235000 | 0.189 | 0.633 | 995 | 90 | -- | -- |
| 99.64 | 0.465 | 3.32 | -0.183 | 0.332 | 5.47 | -- | -- | 0.184 | 0.643 | 994.5 | 105 | -- | -- |
| 119.81 | 0.51 | -150 | 936 | -- | -38400 | -- | 235000 | 0.195 | -- | 995 | 90 | -- | -- |
| 129.3 | 0.533 | 3.33 | -0.18 | 0.326 | -- | 0.0733 | -- | 0.178 | 24 | -- | -- | -- | -- |
| 129.32 | 0.533 | 3.33 | -0.183 | 0.325 | -- | -- | -- | 0.177 | 0.0163 | -- | -- | -- | -- |
| 106.38 | 0.48 | 3.32 | -0.201 | -- | 5.45 | -- | -- | 0.194 | -- | 994.5 | 107 | -- | -- |
| 100.82 | 0.468 | 3.32 | -- | 0.315 | 5.44 | -- | -- | 0.205 | 0.627 | 994.6 | 145 | -- | -- |
| 135.99 | 0.545 | 3.32 | -0.173 | -- | -- | -- | -- | 0.182 | -- | -- | -- | -- | -- |
| 130.2 | 0.534 | 3.32 | -- | 0.323 | -- | -- | -- | 0.198 | 24 | -- | -- | -- | -- |
| 107.44 | 0.483 | 3.31 | -- | -- | 5.42 | -- | -- | 0.216 | -- | 994.6 | 122 | -- | -- |
| 136.77 | 0.546 | 3.32 | -- | -- | -- | -- | -- | 0.201 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance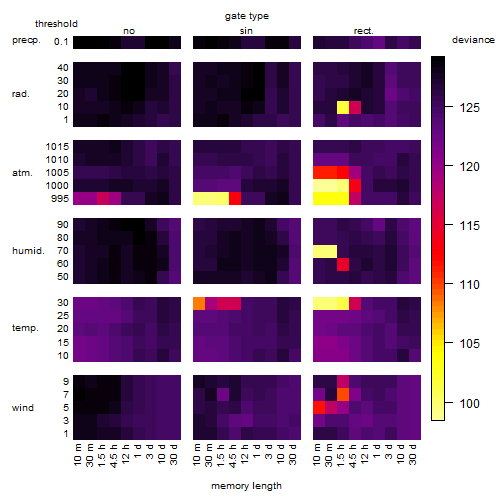
|
Histogram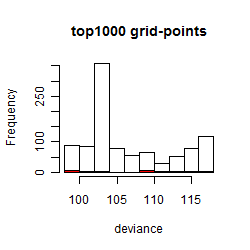 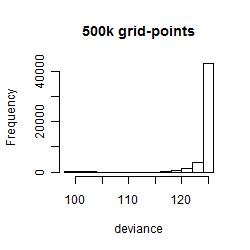
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 98.42 | atmosphere | 1000 | 30 | < th | dose independent | rect. | 16 | 3 |
| 3 | 98.52 | atmosphere | 1000 | 30 | < th | dose dependent | rect. | 16 | 2 |
| 7 | 99.45 | atmosphere | 995 | 10 | > th | dose independent | sin | 8 | NA |
| 23 | 99.52 | temperature | 30 | 10 | > th | dose independent | rect. | 17 | 2 |
| 70 | 99.75 | humidity | 70 | 10 | < th | dose independent | rect. | 18 | 1 |
| 124 | 101.25 | temperature | 30 | 90 | > th | dose dependent | rect. | 16 | 2 |
| 158 | 101.65 | radiation | 10 | 90 | > th | dose dependent | rect. | 17 | 1 |
| 173 | 102.72 | radiation | 10 | 90 | > th | dose independent | rect. | 17 | 1 |
| 400 | 103.75 | atmosphere | 995 | 10 | < th | dose dependent | sin | 8 | NA |
| 504 | 103.92 | atmosphere | 995 | 30 | < th | dose independent | sin | 9 | NA |
| 677 | 108.12 | humidity | 70 | 10 | < th | dose dependent | rect. | 18 | 1 |
| 680 | 108.35 | temperature | 30 | 10 | > th | dose independent | sin | 12 | NA |
| 720 | 109.62 | temperature | 30 | 10 | < th | dose independent | sin | 12 | NA |
| 721 | 109.62 | wind | 7 | 90 | > th | dose independent | rect. | 16 | 1 |
| 722 | 109.67 | wind | 7 | 90 | > th | dose dependent | rect. | 16 | 1 |
| 746 | 111.81 | wind | 5 | 10 | > th | dose dependent | rect. | 17 | 2 |
| 769 | 113.35 | atmosphere | 995 | 270 | > th | dose independent | sin | 4 | NA |
| 839 | 114.54 | humidity | 60 | 90 | < th | dose dependent | rect. | 16 | 1 |
| 851 | 114.82 | atmosphere | 1000 | 270 | < th | dose dependent | rect. | 14 | 1 |
| 879 | 115.84 | wind | 5 | 10 | > th | dose independent | rect. | 17 | 2 |
| 890 | 116.28 | temperature | 30 | 270 | > th | dose dependent | sin | 12 | NA |
| 909 | 116.43 | temperature | 30 | 270 | > th | dose dependent | rect. | 14 | 4 |
| 951 | 116.63 | atmosphere | 995 | 10 | > th | dose independent | rect. | 23 | 23 |