Os12g0640800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Serine/threonine-protein kinase PRP4 homolog (EC 2.7.1.37) (PRP4 pre- mRNA processing factor 4 homolog) (PRP4 kinase).
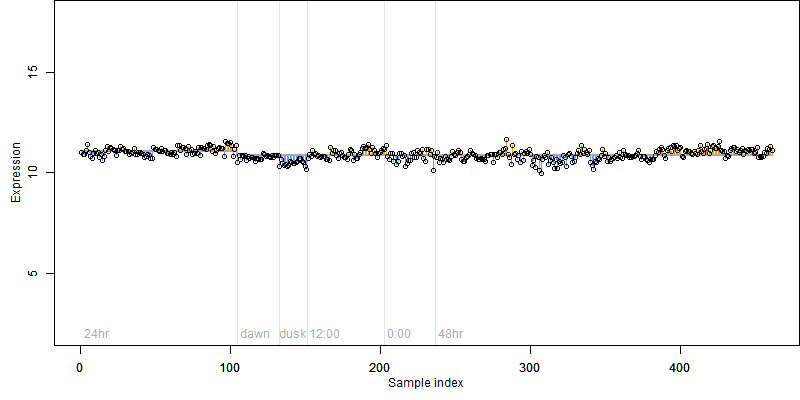

FiT-DB / Search/ Help/ Sample detail
|
Os12g0640800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Serine/threonine-protein kinase PRP4 homolog (EC 2.7.1.37) (PRP4 pre- mRNA processing factor 4 homolog) (PRP4 kinase).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable

Residual plot

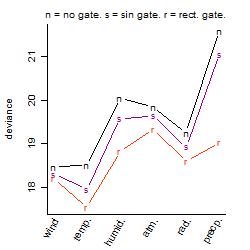
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17.98 | 0.199 | 10.8 | 0.917 | 0.216 | -0.616 | 0.153 | 2.29 | 0.369 | 6.95 | 26.6 | 1239 | -- | -- |
| 18.85 | 0.202 | 10.9 | 0.383 | 0.0886 | -0.499 | -0.0399 | -- | 0.282 | 6.01 | 25.41 | 855 | -- | -- |
| 18.04 | 0.198 | 10.8 | 0.928 | 0.217 | -0.622 | -- | 2.34 | 0.368 | 6.83 | 26.69 | 1238 | -- | -- |
| 18.84 | 0.202 | 10.9 | 0.382 | 0.102 | -0.493 | -- | -- | 0.272 | 6.28 | 25.69 | 887 | -- | -- |
| 19.86 | 0.208 | 10.8 | 1 | -- | -0.766 | -- | 2.93 | 0.389 | -- | 26.7 | 1946 | -- | -- |
| 24.7 | 0.233 | 10.9 | 0.264 | 0.229 | -- | 0.166 | -- | 0.251 | 8.07 | -- | -- | -- | -- |
| 24.78 | 0.233 | 10.9 | 0.268 | 0.229 | -- | -- | -- | 0.251 | 7.97 | -- | -- | -- | -- |
| 18.98 | 0.203 | 10.9 | 0.39 | -- | -0.555 | -- | -- | 0.287 | -- | 25.35 | 770 | -- | -- |
| 22.43 | 0.221 | 10.9 | -- | 0.13 | -0.398 | -- | -- | 0.21 | 6.88 | 27.25 | 885 | -- | -- |
| 27.64 | 0.246 | 10.9 | 0.271 | -- | -- | -- | -- | 0.254 | -- | -- | -- | -- | -- |
| 26.67 | 0.242 | 10.9 | -- | 0.231 | -- | -- | -- | 0.221 | 7.88 | -- | -- | -- | -- |
| 22.75 | 0.222 | 10.9 | -- | -- | -0.492 | -- | -- | 0.222 | -- | 26.3 | 736 | -- | -- |
| 29.57 | 0.254 | 10.9 | -- | -- | -- | -- | -- | 0.224 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance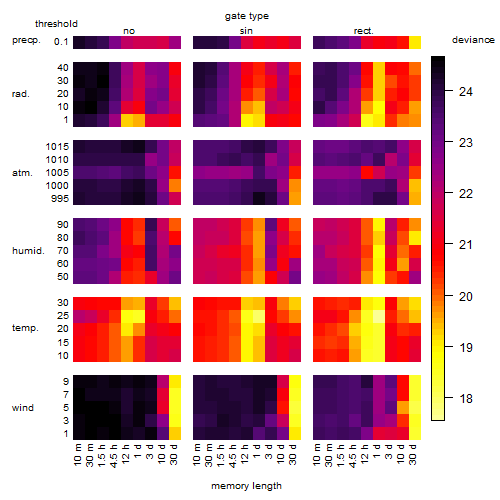
|
Histogram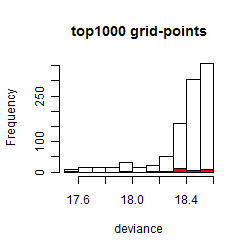 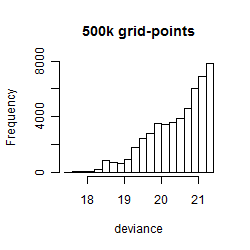
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 17.55 | temperature | 25 | 1440 | > th | dose dependent | rect. | 13 | 6 |
| 2 | 17.56 | temperature | 25 | 1440 | > th | dose dependent | rect. | 17 | 1 |
| 71 | 17.96 | temperature | 25 | 1440 | > th | dose dependent | sin | 18 | NA |
| 127 | 18.20 | wind | 5 | 43200 | > th | dose dependent | rect. | 5 | 16 |
| 169 | 18.30 | wind | 5 | 43200 | > th | dose dependent | sin | 21 | NA |
| 238 | 18.35 | wind | 7 | 43200 | < th | dose independent | rect. | 6 | 15 |
| 246 | 18.35 | wind | 5 | 43200 | > th | dose dependent | rect. | 13 | 19 |
| 253 | 18.36 | wind | 7 | 43200 | < th | dose independent | rect. | 2 | 19 |
| 269 | 18.37 | wind | 7 | 43200 | > th | dose independent | rect. | 6 | 15 |
| 270 | 18.37 | wind | 7 | 43200 | < th | dose independent | rect. | 23 | 22 |
| 272 | 18.37 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 1 |
| 286 | 18.38 | wind | 7 | 43200 | > th | dose independent | rect. | 2 | 19 |
| 289 | 18.38 | wind | 7 | 43200 | > th | dose independent | rect. | 13 | 22 |
| 290 | 18.38 | wind | 7 | 43200 | > th | dose independent | rect. | 23 | 22 |
| 291 | 18.38 | wind | 7 | 43200 | < th | dose independent | rect. | 13 | 22 |
| 315 | 18.39 | wind | 1 | 43200 | > th | dose dependent | rect. | 20 | 1 |
| 343 | 18.41 | wind | 9 | 43200 | < th | dose dependent | rect. | 20 | 1 |
| 522 | 18.47 | wind | 5 | 43200 | > th | dose dependent | no | NA | NA |
| 572 | 18.49 | wind | 5 | 43200 | > th | dose independent | rect. | 5 | 13 |
| 595 | 18.49 | wind | 7 | 43200 | > th | dose independent | rect. | 10 | 23 |
| 603 | 18.49 | temperature | 10 | 720 | > th | dose dependent | rect. | 6 | 18 |
| 634 | 18.50 | temperature | 25 | 1440 | > th | dose dependent | no | NA | NA |
| 686 | 18.51 | wind | 5 | 43200 | > th | dose dependent | rect. | 20 | 23 |
| 687 | 18.52 | wind | 7 | 43200 | < th | dose independent | no | NA | NA |
| 709 | 18.52 | wind | 7 | 43200 | > th | dose independent | rect. | 20 | 23 |
| 737 | 18.53 | wind | 5 | 43200 | > th | dose independent | sin | 23 | NA |
| 739 | 18.53 | wind | 7 | 43200 | > th | dose independent | no | NA | NA |
| 767 | 18.53 | wind | 7 | 43200 | < th | dose independent | rect. | 20 | 23 |
| 808 | 18.54 | temperature | 10 | 1440 | > th | dose dependent | rect. | 14 | 5 |
| 879 | 18.56 | wind | 5 | 43200 | < th | dose independent | sin | 0 | NA |
| 890 | 18.56 | wind | 7 | 43200 | < th | dose independent | rect. | 10 | 17 |
| 937 | 18.57 | wind | 7 | 43200 | < th | dose independent | sin | 17 | NA |