Os12g0571700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Hypothetical protein.
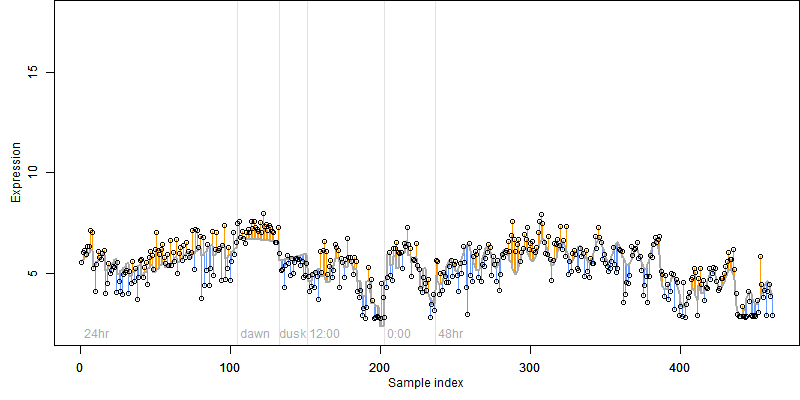
FiT-DB / Search/ Help/ Sample detail
|
Os12g0571700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
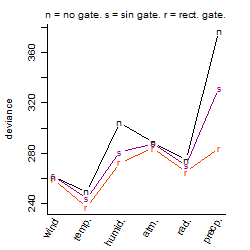
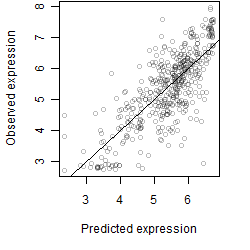
Dependence on each variable

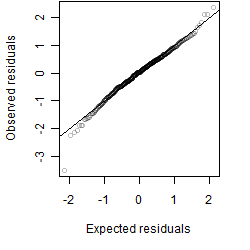
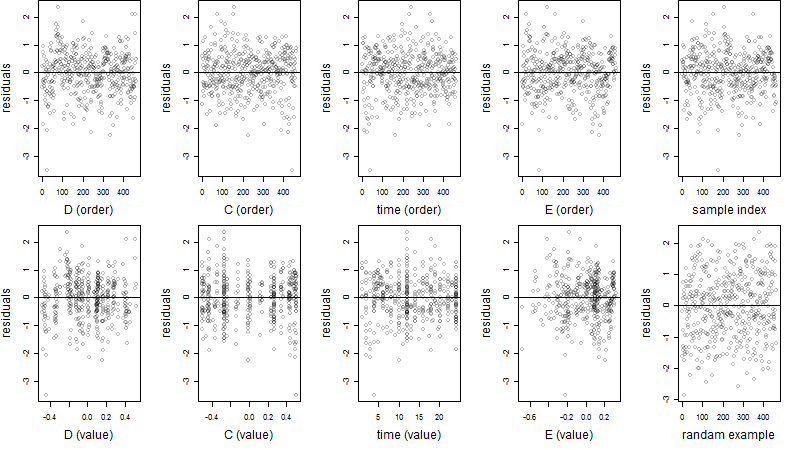
Residual plot
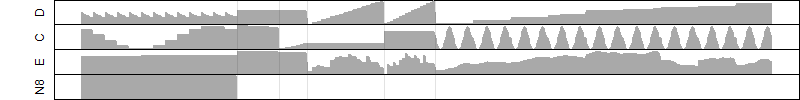
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 248.77 | 0.741 | 5.56 | -3.39 | 0.974 | 3.54 | 1.19 | -2.76 | -0.673 | 3.64 | 11.5 | 1374 | -- | -- |
| 254.95 | 0.744 | 5.49 | -2.79 | 0.926 | 3.58 | 1.13 | -- | -0.513 | 3.77 | 13.22 | 1310 | -- | -- |
| 252.91 | 0.741 | 5.55 | -3.33 | 0.96 | 3.55 | -- | -2.41 | -0.658 | 3.75 | 11.2 | 1346 | -- | -- |
| 257.75 | 0.748 | 5.48 | -2.8 | 0.919 | 3.56 | -- | -- | -0.519 | 3.91 | 12.59 | 1306 | -- | -- |
| 295.8 | 0.801 | 5.56 | -3.29 | -- | 3.85 | -- | -2.21 | -0.664 | -- | 7.934 | 1218 | -- | -- |
| 463.81 | 1.01 | 5.34 | -1.68 | 1.21 | -- | 1.12 | -- | 0.127 | 3.42 | -- | -- | -- | -- |
| 467.75 | 1.01 | 5.34 | -1.7 | 1.21 | -- | -- | -- | 0.121 | 3.45 | -- | -- | -- | -- |
| 299.63 | 0.806 | 5.5 | -2.82 | -- | 3.84 | -- | -- | -0.536 | -- | 13.33 | 1217 | -- | -- |
| 427.05 | 0.962 | 5.38 | -- | 0.954 | 1.9 | -- | -- | -0.038 | 4.29 | 21.15 | 1163 | -- | -- |
| 550.39 | 1.1 | 5.34 | -1.65 | -- | -- | -- | -- | 0.153 | -- | -- | -- | -- | -- |
| 544.05 | 1.09 | 5.3 | -- | 1.18 | -- | -- | -- | 0.31 | 3.45 | -- | -- | -- | -- |
| 472.26 | 1.01 | 5.38 | -- | -- | 2.06 | -- | -- | -0.0426 | -- | 21.3 | 1184 | -- | -- |
| 622.23 | 1.16 | 5.3 | -- | -- | -- | -- | -- | 0.336 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance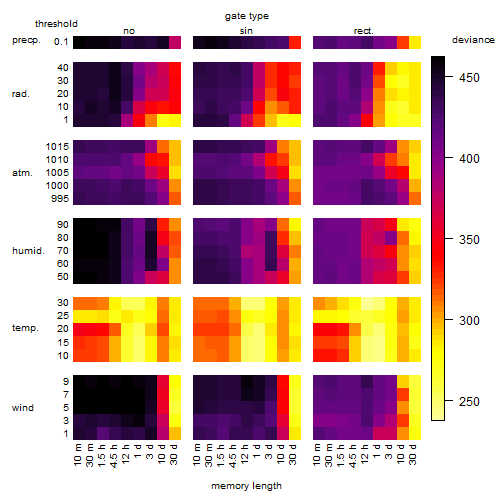
|
Histogram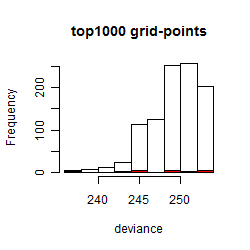 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 237.20 | temperature | 30 | 720 | < th | dose dependent | rect. | 10 | 18 |
| 27 | 242.38 | temperature | 10 | 1440 | > th | dose dependent | rect. | 16 | 7 |
| 52 | 244.26 | temperature | 10 | 1440 | > th | dose dependent | sin | 16 | NA |
| 64 | 244.48 | temperature | 10 | 1440 | > th | dose dependent | rect. | 13 | 14 |
| 95 | 245.02 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 1 |
| 111 | 245.22 | temperature | 30 | 720 | < th | dose dependent | sin | 15 | NA |
| 113 | 245.24 | temperature | 30 | 1440 | < th | dose dependent | rect. | 16 | 7 |
| 399 | 249.12 | temperature | 10 | 1440 | > th | dose dependent | rect. | 2 | 22 |
| 473 | 249.58 | temperature | 10 | 1440 | > th | dose dependent | no | NA | NA |
| 528 | 249.97 | temperature | 10 | 1440 | > th | dose dependent | rect. | 21 | 23 |
| 531 | 249.98 | temperature | 10 | 1440 | > th | dose dependent | rect. | 16 | 23 |
| 632 | 250.64 | temperature | 30 | 720 | < th | dose dependent | rect. | 1 | 23 |
| 878 | 252.61 | temperature | 30 | 1440 | < th | dose dependent | no | NA | NA |
| 881 | 252.69 | temperature | 30 | 1440 | < th | dose dependent | rect. | 16 | 23 |
| 936 | 253.21 | temperature | 30 | 1440 | < th | dose dependent | rect. | 21 | 23 |