FiT-DB /
Search/
Help/
Sample detail
Notice!
This gene did NOT pass the analysis criteria
(log2(expression) > 5 in more than 80% of samples &
normality of residuals was not rejected).
The modeling results may be unreliable.
|
Os12g0434400
|
blastp(Os)/
blastp(At)/
coex///
RAP/
RiceXPro/
SALAD/
ATTED-II
|
|
Description : Similar to Cytosol aminopeptidase (EC 3.4.11.1) (Leucine aminopeptidase) (LAP) (Leucyl aminopeptidase) (Proline aminopeptidase) (EC 3.4.11.5) (Prolyl aminopeptidase).


|
log2(Expression) ~
Norm(μ, σ2)
μ = α
+ β1D
+ β2C
+ β3E
+ β4D*C
+ β5D*E
+ γ1N8
| par. |
|
value |
(S.E.) |
|
|
|
|
|
|
| α |
: |
3.68 |
(0.054) |
R2 |
: |
0.468 |
|
|
|
| β1 |
: |
3.35 |
(0.213) |
R2dD |
: |
0.361 |
R2D |
: |
-0.0341 |
| β2 |
: |
0.801 |
(0.129) |
R2dC |
: |
0.0464 |
R2C |
: |
-0.0373 |
| β3 |
: |
-3.36 |
(0.205) |
R2dE |
: |
0.414 |
R2E |
: |
-0.0838 |
| β4 |
: |
-- |
(--) |
R2dD*C |
: |
-- |
R2D*C |
: |
-- |
| β5 |
: |
-- |
(--) |
R2dD*E |
: |
-- |
R2D*E |
: |
-- |
| γ1 |
: |
1.38 |
(0.122) |
R2dN8 |
: |
0.233 |
R2N8 |
: |
-0.0651 |
| σ |
: |
0.993 |
|
|
|
|
|
|
|
| deviance |
: |
454.26 |
|
|
|
|
|
|
|
| __ |
|
|
|
|
|
|
|
|
|
|
_____
|
| C |
|
|
| peak time of C |
: |
1.6 |
| E |
|
|
| wheather |
: |
temperature |
| threshold |
: |
16.97 |
| memory length |
: |
2776 |
| response mode |
: |
> th |
| dose dependency |
: |
dose dependent |
| G |
|
|
| type of G |
: |
no |
| peak or start time of G |
: |
-- |
| open length of G |
: |
-- |
|
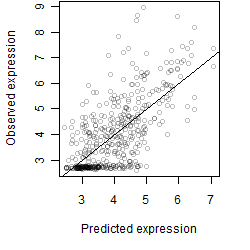 __
__
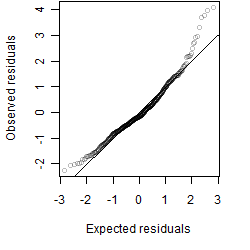
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
| 452.14 |
0.999 |
3.74 |
3 |
0.693 |
-3.68 |
0.341 |
-1.95 |
1.27 |
1.61 |
14.93 |
2991 |
-- |
-- |
| 454.72 |
0.993 |
3.65 |
3.37 |
0.699 |
-3.42 |
0.221 |
-- |
1.37 |
1.71 |
16.33 |
2842 |
-- |
-- |
| 452.56 |
0.991 |
3.73 |
3.03 |
0.681 |
-3.57 |
-- |
-1.62 |
1.28 |
1.56 |
15.8 |
2993 |
-- |
-- |
| 454.26 |
0.993 |
3.68 |
3.35 |
0.801 |
-3.36 |
-- |
-- |
1.38 |
1.6 |
16.97 |
2776 |
-- |
-- |
| 476.18 |
1.02 |
3.73 |
3.28 |
-- |
-3.6 |
-- |
-1.67 |
1.28 |
-- |
15.69 |
4660 |
-- |
-- |
| 720.84 |
1.26 |
3.84 |
1.95 |
0.489 |
-- |
0.213 |
-- |
0.649 |
1.27 |
-- |
-- |
-- |
-- |
| 721.02 |
1.26 |
3.84 |
1.94 |
0.488 |
-- |
-- |
-- |
0.648 |
1.21 |
-- |
-- |
-- |
-- |
| 478.14 |
1.02 |
3.67 |
3.64 |
-- |
-3.45 |
-- |
-- |
1.39 |
-- |
16.9 |
4666 |
-- |
-- |
| 692.5 |
1.23 |
3.77 |
-- |
0.777 |
-2.62 |
-- |
-- |
0.903 |
1.81 |
16.85 |
1244 |
-- |
-- |
| 735.77 |
1.27 |
3.83 |
1.96 |
-- |
-- |
-- |
-- |
0.658 |
-- |
-- |
-- |
-- |
-- |
| 820.72 |
1.34 |
3.89 |
-- |
0.522 |
-- |
-- |
-- |
0.431 |
1.36 |
-- |
-- |
-- |
-- |
| 692.31 |
1.23 |
3.8 |
-- |
-- |
-1.99 |
-- |
-- |
0.81 |
-- |
21.88 |
1657 |
-- |
-- |
| 837.49 |
1.35 |
3.88 |
-- |
-- |
-- |
-- |
-- |
0.44 |
-- |
-- |
-- |
-- |
-- |
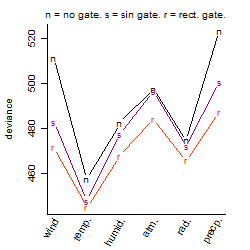
Results of the grid search
Summarized heatmap of deviance

|
Histogram


|
Local optima within top1000 grid-points
| rank |
deviance |
wheather |
threshold |
memory length |
response mode |
dose dependency |
type of G |
peak or start time of G |
open length of G |
| 1 |
444.79 |
temperature |
15 |
4320 |
> th |
dose dependent |
rect. |
24 |
4 |
| 3 |
444.87 |
temperature |
15 |
4320 |
> th |
dose dependent |
rect. |
24 |
6 |
| 7 |
445.03 |
temperature |
15 |
4320 |
> th |
dose dependent |
rect. |
3 |
1 |
| 23 |
445.72 |
temperature |
30 |
4320 |
< th |
dose dependent |
rect. |
24 |
4 |
| 24 |
445.76 |
temperature |
30 |
4320 |
< th |
dose dependent |
rect. |
23 |
8 |
| 82 |
446.48 |
temperature |
30 |
4320 |
< th |
dose dependent |
rect. |
3 |
1 |
| 160 |
447.54 |
temperature |
15 |
4320 |
> th |
dose dependent |
sin |
10 |
NA |
| 195 |
448.02 |
temperature |
30 |
4320 |
< th |
dose dependent |
sin |
10 |
NA |
| 218 |
448.30 |
temperature |
15 |
4320 |
> th |
dose dependent |
rect. |
5 |
1 |
| 303 |
449.44 |
temperature |
30 |
4320 |
< th |
dose dependent |
rect. |
5 |
1 |
| 978 |
457.02 |
temperature |
15 |
4320 |
> th |
dose dependent |
no |
NA |
NA |



 __
__





