Os12g0177800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein kinase domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os12g0177800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein kinase domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

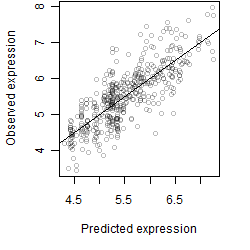
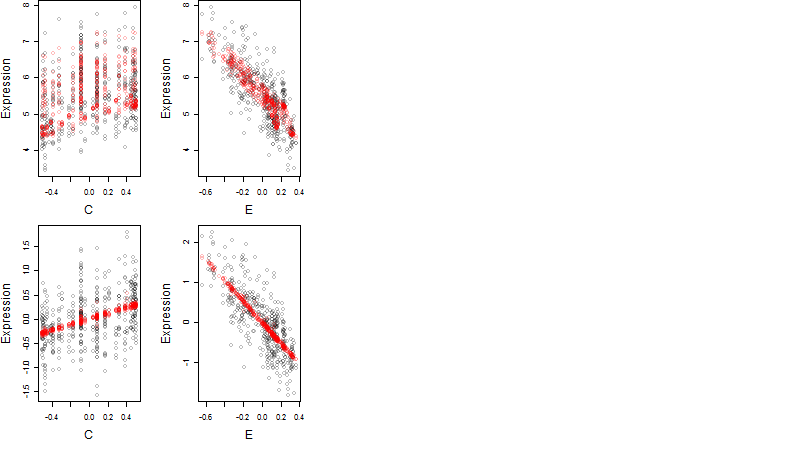
Dependence on each variable
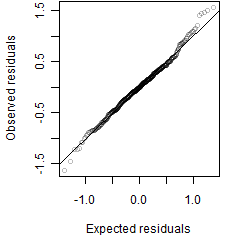
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 105 | 0.481 | 5.47 | 0.429 | 0.677 | -2.09 | 0.555 | 2.4 | -0.0654 | 6.48 | 20.36 | 1649 | -- | -- |
| 108.78 | 0.486 | 5.55 | -0.122 | 0.583 | -2.56 | 0.183 | -- | -0.232 | 5.45 | 10.29 | 2083 | -- | -- |
| 105.75 | 0.479 | 5.47 | 0.422 | 0.679 | -2.09 | -- | 2.35 | -0.0712 | 6.39 | 20.3 | 1649 | -- | -- |
| 108.85 | 0.486 | 5.55 | -0.104 | 0.59 | -2.56 | -- | -- | -0.225 | 5.43 | 10.08 | 2063 | -- | -- |
| 121.64 | 0.514 | 5.48 | 0.478 | -- | -2.24 | -- | 2.45 | -0.0369 | -- | 20.73 | 1924 | -- | -- |
| 223.51 | 0.701 | 5.65 | -0.996 | 0.749 | -- | 0.501 | -- | -0.717 | 6.63 | -- | -- | -- | -- |
| 224.13 | 0.701 | 5.66 | -0.988 | 0.751 | -- | -- | -- | -0.719 | 6.56 | -- | -- | -- | -- |
| 126 | 0.523 | 5.56 | -0.148 | -- | -2.25 | -- | -- | -0.205 | -- | 18.82 | 1964 | -- | -- |
| 109.09 | 0.486 | 5.55 | -- | 0.593 | -2.6 | -- | -- | -0.207 | 5.34 | 10.41 | 2085 | -- | -- |
| 253.51 | 0.744 | 5.67 | -0.97 | -- | -- | -- | -- | -0.704 | -- | -- | -- | -- | -- |
| 249.87 | 0.739 | 5.63 | -- | 0.739 | -- | -- | -- | -0.609 | 6.64 | -- | -- | -- | -- |
| 126.12 | 0.523 | 5.55 | -- | -- | -2.72 | -- | -- | -0.183 | -- | 14.6 | 1977 | -- | -- |
| 278.34 | 0.779 | 5.65 | -- | -- | -- | -- | -- | -0.596 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance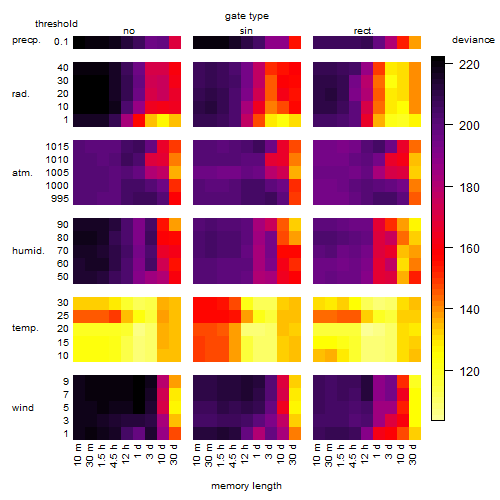
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 103.55 | temperature | 20 | 720 | > th | dose dependent | rect. | 20 | 18 |
| 8 | 105.74 | temperature | 20 | 1440 | > th | dose dependent | rect. | 16 | 22 |
| 13 | 105.94 | temperature | 20 | 1440 | > th | dose dependent | rect. | 11 | 22 |
| 21 | 106.13 | temperature | 20 | 1440 | > th | dose dependent | rect. | 24 | 22 |
| 32 | 106.25 | temperature | 20 | 1440 | > th | dose dependent | no | NA | NA |
| 44 | 106.35 | temperature | 20 | 1440 | > th | dose dependent | rect. | 20 | 23 |
| 47 | 106.36 | temperature | 20 | 1440 | > th | dose dependent | rect. | 6 | 23 |
| 49 | 106.37 | temperature | 20 | 1440 | > th | dose dependent | rect. | 3 | 23 |
| 50 | 106.39 | temperature | 20 | 1440 | > th | dose dependent | rect. | 15 | 18 |
| 148 | 107.33 | temperature | 20 | 1440 | > th | dose dependent | sin | 15 | NA |
| 151 | 107.40 | temperature | 20 | 1440 | > th | dose dependent | sin | 5 | NA |
| 152 | 107.40 | temperature | 20 | 1440 | > th | dose dependent | rect. | 24 | 15 |
| 155 | 107.42 | temperature | 20 | 1440 | > th | dose dependent | rect. | 1 | 13 |
| 200 | 107.83 | temperature | 20 | 1440 | > th | dose dependent | rect. | 15 | 14 |
| 207 | 107.88 | temperature | 20 | 1440 | > th | dose dependent | rect. | 17 | 5 |
| 208 | 107.89 | temperature | 30 | 1440 | < th | dose dependent | rect. | 17 | 4 |
| 218 | 108.02 | temperature | 30 | 1440 | < th | dose dependent | rect. | 17 | 2 |
| 509 | 109.24 | temperature | 20 | 1440 | > th | dose dependent | rect. | 18 | 1 |
| 520 | 109.27 | temperature | 10 | 4320 | > th | dose dependent | rect. | 17 | 1 |
| 636 | 109.70 | temperature | 20 | 1440 | > th | dose dependent | rect. | 4 | 5 |
| 643 | 109.73 | temperature | 30 | 1440 | < th | dose dependent | rect. | 16 | 18 |