Os12g0137700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Sulfotransferase family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os12g0137700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Sulfotransferase family protein.
|
|
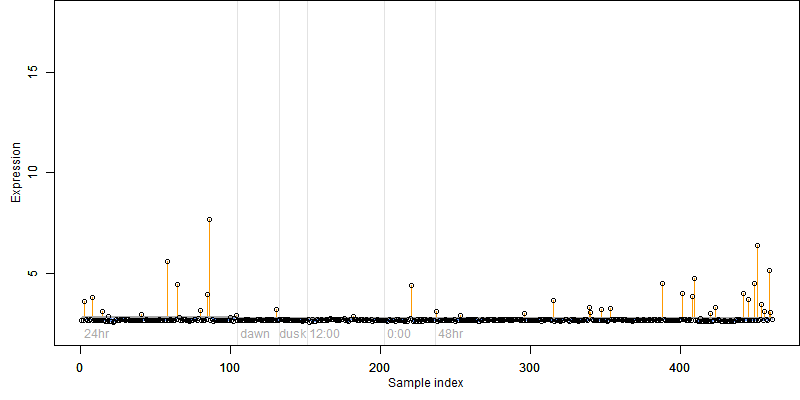
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
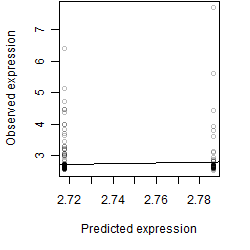
Dependence on each variable

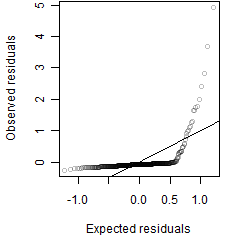
Residual plot

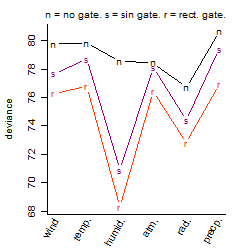
Process of the parameter reduction
(fixed parameters. wheather = radiation,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 76.73 | 0.412 | 2.71 | 0.256 | 0.134 | 0.284 | 0.383 | 2.51 | 0.0913 | 3.62 | 40.01 | 272 | -- | -- |
| 80.8 | 0.419 | 2.71 | 0.297 | 0.172 | 0.365 | 0.0431 | -- | 0.0745 | 3.56 | 42.4 | 259 | -- | -- |
| 77.13 | 0.409 | 2.72 | 0.263 | 0.142 | 0.3 | -- | 2.43 | 0.0896 | 3.46 | 40.63 | 264 | -- | -- |
| 80.52 | 0.418 | 2.72 | 0.221 | 0.147 | 0.342 | -- | -- | 0.068 | 3.25 | 42.42 | 259 | -- | -- |
| 77.81 | 0.411 | 2.72 | 0.258 | -- | 0.171 | -- | 2.16 | 0.0998 | -- | 40.73 | 262 | -- | -- |
| 81.5 | 0.423 | 2.71 | 0.216 | 0.0527 | -- | -0.179 | -- | 0.0909 | 3.03 | -- | -- | -- | -- |
| 81.58 | 0.423 | 2.71 | 0.22 | 0.0577 | -- | -- | -- | 0.0918 | 4.42 | -- | -- | -- | -- |
| 81.33 | 0.42 | 2.71 | 0.223 | -- | 0.177 | -- | -- | 0.0837 | -- | 43.31 | 253 | -- | -- |
| 81.8 | 0.421 | 2.72 | -- | 0.152 | 0.345 | -- | -- | 0.0589 | 3.16 | 42.12 | 263 | -- | -- |
| 81.76 | 0.423 | 2.71 | 0.222 | -- | -- | -- | -- | 0.0934 | -- | -- | -- | -- | -- |
| 82.86 | 0.426 | 2.72 | -- | 0.0618 | -- | -- | -- | 0.0673 | 4.34 | -- | -- | -- | -- |
| 82.64 | 0.423 | 2.72 | -- | -- | 0.175 | -- | -- | 0.0588 | -- | 42.42 | 259 | -- | -- |
| 83.06 | 0.425 | 2.72 | -- | -- | -- | -- | -- | 0.0687 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance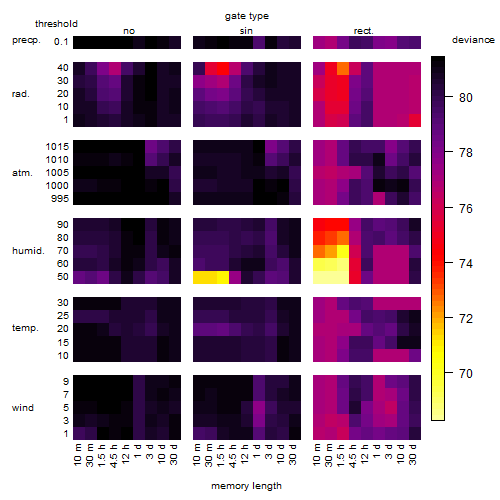
|
Histogram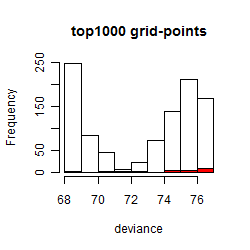 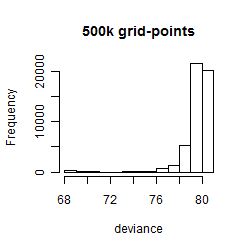
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 68.27 | humidity | 50 | 90 | < th | dose dependent | rect. | 12 | 2 |
| 2 | 68.28 | humidity | 50 | 90 | < th | dose independent | rect. | 13 | 1 |
| 378 | 70.89 | humidity | 50 | 90 | < th | dose dependent | sin | 7 | NA |
| 381 | 71.30 | humidity | 50 | 10 | < th | dose dependent | sin | 12 | NA |
| 383 | 71.33 | humidity | 50 | 10 | < th | dose independent | sin | 13 | NA |
| 388 | 72.03 | humidity | 50 | 10 | > th | dose independent | sin | 13 | NA |
| 399 | 72.78 | radiation | 40 | 90 | > th | dose independent | rect. | 12 | 2 |
| 527 | 74.35 | radiation | 40 | 90 | > th | dose independent | sin | 14 | NA |
| 571 | 74.57 | humidity | 50 | 90 | < th | dose independent | sin | 7 | NA |
| 614 | 74.92 | radiation | 20 | 90 | > th | dose dependent | rect. | 12 | 2 |
| 691 | 75.34 | radiation | 30 | 90 | > th | dose dependent | rect. | 12 | 21 |
| 700 | 75.42 | radiation | 1 | 43200 | < th | dose dependent | rect. | 3 | 1 |
| 792 | 75.91 | radiation | 40 | 90 | < th | dose independent | sin | 14 | NA |
| 864 | 76.19 | radiation | 40 | 270 | < th | dose independent | rect. | 6 | 11 |
| 884 | 76.24 | radiation | 40 | 270 | > th | dose independent | rect. | 9 | 5 |
| 887 | 76.25 | wind | 1 | 90 | < th | dose dependent | rect. | 4 | 1 |
| 982 | 76.38 | wind | 3 | 30 | > th | dose independent | rect. | 13 | 1 |
| 983 | 76.38 | wind | 5 | 4320 | > th | dose dependent | rect. | 21 | 4 |
| 985 | 76.40 | wind | 5 | 4320 | > th | dose independent | rect. | 21 | 4 |
| 987 | 76.41 | atmosphere | 1005 | 30 | > th | dose dependent | rect. | 5 | 1 |
| 989 | 76.43 | wind | 5 | 4320 | < th | dose independent | rect. | 21 | 4 |