Os11g0699100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Disease resistance protein family protein.
FiT-DB / Search/ Help/ Sample detail
|
Os11g0699100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Disease resistance protein family protein.
|
|
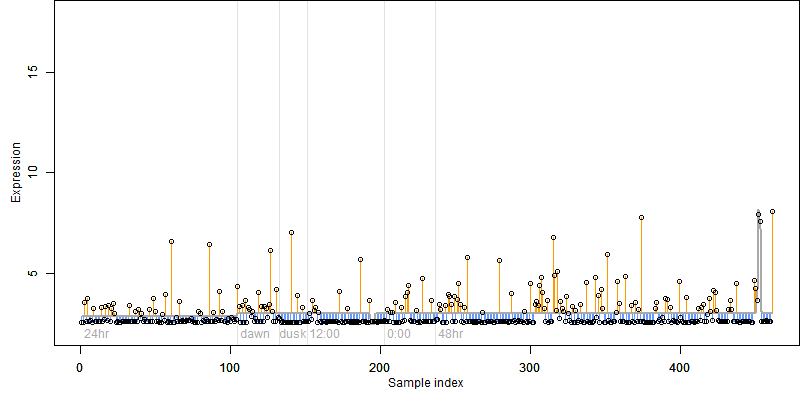
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

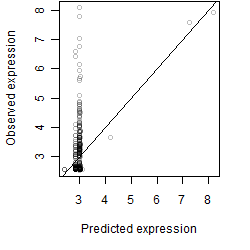
Dependence on each variable

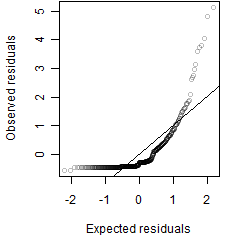
Residual plot
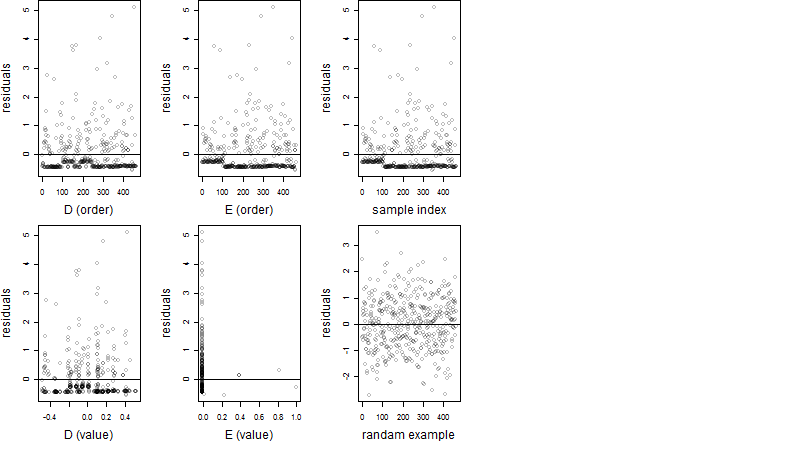
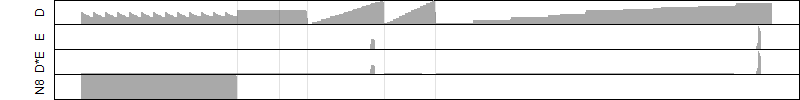
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 271.37 | 0.774 | 2.32 | 1.66 | 0.0451 | -97 | 1.12 | 248 | -0.163 | 8.74 | 47.01 | 279 | -- | -- |
| 313.91 | 0.832 | 3.01 | 0.108 | 0.000572 | -0.676 | 0.951 | -- | -0.17 | 12 | 45.26 | 199 | -- | -- |
| 272.33 | 0.769 | 2.31 | 1.7 | 0.148 | -97.4 | -- | 250 | -0.155 | 5.09 | 45 | 265 | -- | -- |
| 316.41 | 0.828 | 3.01 | 0.196 | 0.0395 | -0.392 | -- | -- | -0.17 | 0.918 | 37.99 | 4126 | -- | -- |
| 273.43 | 0.77 | 2.3 | 1.74 | -- | -97.8 | -- | 250 | -0.163 | -- | 44.77 | 266 | -- | -- |
| 315.2 | 0.832 | 3.01 | 0.0831 | 0.0162 | -- | -0.866 | -- | -0.167 | 0.0136 | -- | -- | -- | -- |
| 318.35 | 0.836 | 3.01 | 0.116 | 0.0496 | -- | -- | -- | -0.164 | 4.47 | -- | -- | -- | -- |
| 316.48 | 0.829 | 3.01 | 0.199 | -- | -0.402 | -- | -- | -0.17 | -- | 37.13 | 4121 | -- | -- |
| 317.29 | 0.83 | 3.02 | -- | 0.0491 | -0.327 | -- | -- | -0.198 | 3.39 | 36 | 4038 | -- | -- |
| 318.48 | 0.834 | 3.01 | 0.118 | -- | -- | -- | -- | -0.163 | -- | -- | -- | -- | -- |
| 318.7 | 0.835 | 3.01 | -- | 0.0518 | -- | -- | -- | -0.177 | 4.41 | -- | -- | -- | -- |
| 317.44 | 0.83 | 3.02 | -- | -- | -0.324 | -- | -- | -0.189 | -- | 37.39 | 4076 | -- | -- |
| 318.85 | 0.833 | 3.01 | -- | -- | -- | -- | -- | -0.176 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 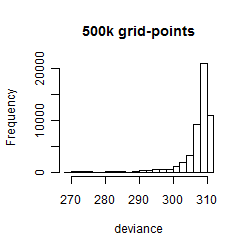
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 269.05 | wind | 1 | 10 | < th | dose independent | rect. | 14 | 4 |
| 5 | 270.18 | humidity | 50 | 270 | < th | dose dependent | rect. | 14 | 2 |
| 24 | 270.29 | humidity | 50 | 270 | < th | dose dependent | sin | 13 | NA |
| 25 | 270.50 | humidity | 50 | 270 | < th | dose independent | rect. | 14 | 2 |
| 26 | 270.51 | humidity | 50 | 270 | < th | dose independent | rect. | 14 | 4 |
| 64 | 270.85 | humidity | 50 | 270 | < th | dose independent | sin | 13 | NA |
| 115 | 273.21 | wind | 1 | 10 | < th | dose dependent | rect. | 14 | 4 |
| 144 | 273.78 | humidity | 50 | 270 | > th | dose independent | sin | 13 | NA |
| 324 | 281.65 | humidity | 50 | 270 | < th | dose dependent | no | NA | NA |
| 447 | 282.67 | radiation | 30 | 90 | > th | dose independent | rect. | 8 | 1 |
| 511 | 284.41 | radiation | 20 | 90 | > th | dose dependent | rect. | 8 | 1 |
| 602 | 285.66 | humidity | 50 | 270 | < th | dose independent | no | NA | NA |
| 724 | 285.75 | humidity | 50 | 270 | < th | dose dependent | sin | 7 | NA |
| 732 | 287.14 | humidity | 50 | 270 | > th | dose independent | rect. | 7 | 23 |
| 754 | 287.97 | radiation | 40 | 10 | > th | dose dependent | rect. | 10 | 1 |
| 756 | 288.19 | wind | 1 | 90 | < th | dose dependent | rect. | 8 | 2 |
| 780 | 288.76 | humidity | 50 | 270 | > th | dose independent | rect. | 9 | 12 |
| 806 | 289.71 | wind | 3 | 90 | < th | dose independent | rect. | 15 | 3 |
| 807 | 289.86 | humidity | 50 | 270 | > th | dose independent | no | NA | NA |
| 808 | 289.99 | humidity | 70 | 10 | < th | dose independent | rect. | 8 | 2 |
| 831 | 290.48 | radiation | 40 | 30 | > th | dose independent | rect. | 14 | 21 |
| 906 | 291.08 | humidity | 50 | 10 | < th | dose dependent | rect. | 15 | 3 |