Os11g0519100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.
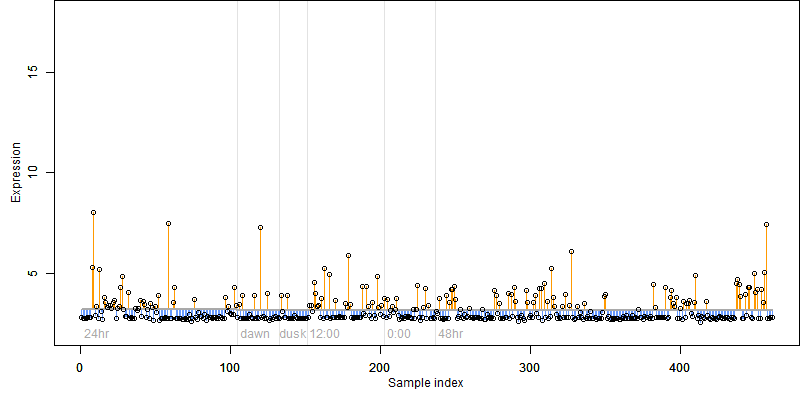

FiT-DB / Search/ Help/ Sample detail
|
Os11g0519100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
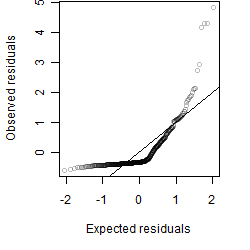
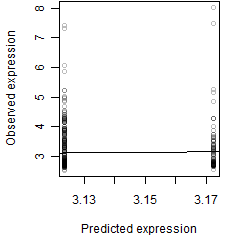 __
__
Dependence on each variable

Residual plot

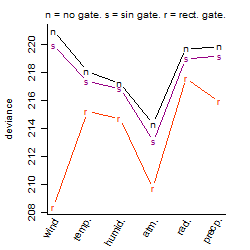
Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 212.96 | 0.686 | 3.14 | -0.0708 | 0.271 | 1.78 | -0.509 | 4.38 | 0.0757 | 11.3 | 1016 | 4109 | -- | -- |
| 223.12 | 0.696 | 3.1 | 0.137 | 0.267 | 0.392 | -0.432 | -- | 0.115 | 11.2 | 1014 | 4983 | -- | -- |
| 213.59 | 0.681 | 3.13 | -0.0609 | 0.312 | 2.57 | -- | 5.68 | 0.114 | 10.6 | 1016 | 4071 | -- | -- |
| 222.47 | 0.695 | 3.1 | 0.243 | 0.245 | 0.393 | -- | -- | 0.154 | 11.2 | 1013 | 15180 | -- | -- |
| 217.88 | 0.687 | 3.16 | -0.0505 | -- | 2.87 | -- | 6.2 | 0.0722 | -- | 1016 | 4063 | -- | -- |
| 227.25 | 0.707 | 3.12 | 0.0666 | 0.255 | -- | -0.564 | -- | 0.0576 | 11.4 | -- | -- | -- | -- |
| 228.62 | 0.708 | 3.12 | 0.0434 | 0.258 | -- | -- | -- | 0.0552 | 11.3 | -- | -- | -- | -- |
| 226.51 | 0.701 | 3.1 | 0.247 | -- | 0.403 | -- | -- | 0.162 | -- | 1013 | 15186 | -- | -- |
| 223.89 | 0.697 | 3.1 | -- | 0.259 | 0.339 | -- | -- | 0.121 | 11.2 | 1013 | 15234 | -- | -- |
| 232.77 | 0.713 | 3.12 | 0.037 | -- | -- | -- | -- | 0.0528 | -- | -- | -- | -- | -- |
| 228.67 | 0.707 | 3.12 | -- | 0.257 | -- | -- | -- | 0.0504 | 11.3 | -- | -- | -- | -- |
| 227.87 | 0.703 | 3.11 | -- | -- | 0.324 | -- | -- | 0.12 | -- | 1013 | 15224 | -- | -- |
| 232.81 | 0.712 | 3.12 | -- | -- | -- | -- | -- | 0.0487 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance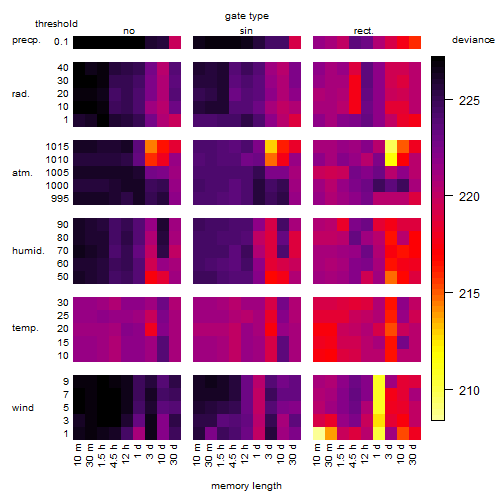
|
Histogram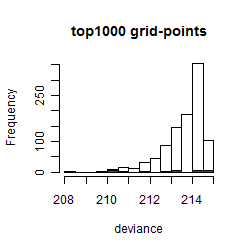 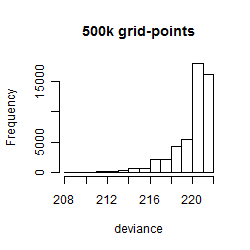
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 208.37 | wind | 1 | 10 | < th | dose dependent | rect. | 1 | 1 |
| 2 | 209.72 | atmosphere | 1015 | 4320 | > th | dose dependent | rect. | 3 | 1 |
| 4 | 210.13 | atmosphere | 1015 | 4320 | > th | dose dependent | rect. | 2 | 3 |
| 5 | 210.14 | wind | 1 | 1440 | > th | dose dependent | rect. | 2 | 1 |
| 7 | 210.23 | atmosphere | 1015 | 4320 | > th | dose independent | rect. | 3 | 1 |
| 13 | 210.50 | atmosphere | 1015 | 4320 | < th | dose independent | rect. | 3 | 1 |
| 21 | 210.75 | wind | 7 | 1440 | < th | dose dependent | rect. | 2 | 1 |
| 51 | 211.74 | atmosphere | 1015 | 4320 | < th | dose independent | rect. | 5 | 1 |
| 156 | 212.77 | wind | 3 | 1440 | < th | dose independent | rect. | 2 | 1 |
| 167 | 212.84 | wind | 3 | 1440 | > th | dose independent | rect. | 2 | 1 |
| 215 | 213.05 | atmosphere | 1015 | 4320 | > th | dose independent | sin | 5 | NA |
| 226 | 213.09 | atmosphere | 1015 | 4320 | < th | dose independent | sin | 5 | NA |
| 240 | 213.14 | atmosphere | 1015 | 4320 | > th | dose dependent | sin | 6 | NA |
| 280 | 213.27 | wind | 1 | 10 | < th | dose independent | rect. | 1 | 1 |
| 746 | 214.29 | atmosphere | 1015 | 4320 | > th | dose independent | no | NA | NA |
| 774 | 214.33 | atmosphere | 1015 | 4320 | < th | dose independent | no | NA | NA |
| 798 | 214.36 | atmosphere | 1015 | 4320 | > th | dose independent | rect. | 9 | 23 |
| 819 | 214.40 | atmosphere | 1015 | 4320 | < th | dose independent | rect. | 9 | 23 |
| 837 | 214.42 | atmosphere | 1015 | 4320 | > th | dose independent | rect. | 7 | 23 |
| 868 | 214.46 | atmosphere | 1015 | 4320 | < th | dose independent | rect. | 7 | 23 |
| 880 | 214.48 | atmosphere | 1015 | 4320 | > th | dose independent | rect. | 5 | 23 |
| 900 | 214.51 | atmosphere | 1015 | 4320 | < th | dose independent | rect. | 5 | 23 |
| 920 | 214.54 | atmosphere | 1015 | 4320 | > th | dose dependent | no | NA | NA |
| 982 | 214.67 | atmosphere | 1015 | 4320 | > th | dose dependent | rect. | 9 | 23 |
| 996 | 214.70 | atmosphere | 1015 | 4320 | > th | dose dependent | rect. | 5 | 23 |