Os11g0270500
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Disease resistance protein family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os11g0270500 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Disease resistance protein family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

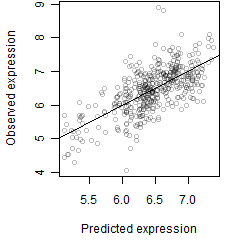 __
__

Dependence on each variable

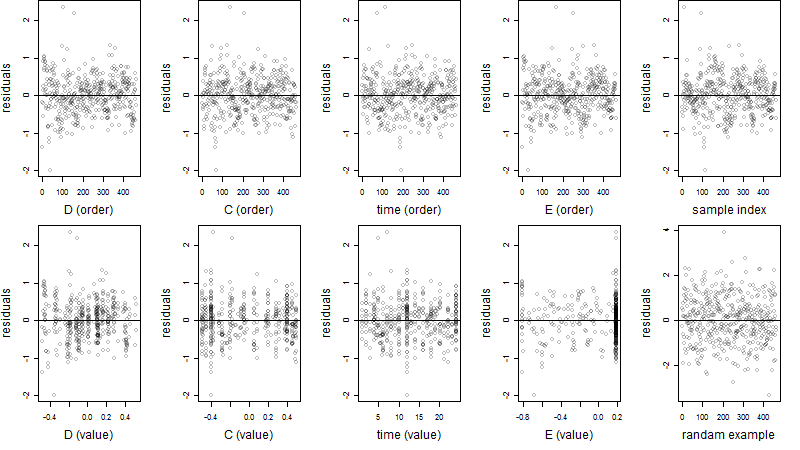
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 109.7 | 0.492 | 6.43 | -0.914 | 0.746 | 0.808 | -1.15 | 1.17 | 0.0828 | 21.9 | 21.4 | 789 | -- | -- |
| 113.75 | 0.497 | 6.45 | -1.2 | 0.741 | 0.722 | -0.745 | -- | 0.0678 | 21.6 | 21.7 | 824 | -- | -- |
| 113.95 | 0.497 | 6.44 | -0.968 | 0.745 | 0.794 | -- | 0.803 | 0.0749 | 21.5 | 21.4 | 857 | -- | -- |
| 115.27 | 0.5 | 6.47 | -1.18 | 0.766 | 0.75 | -- | -- | 0.0171 | 21.6 | 21.5 | 784 | -- | -- |
| 146.1 | 0.563 | 6.44 | -1.05 | -- | 1.02 | -- | 0.774 | -0.00221 | -- | 21.5 | 794 | -- | -- |
| 133.07 | 0.541 | 6.42 | -0.938 | 0.898 | -- | -0.758 | -- | 0.226 | 21.8 | -- | -- | -- | -- |
| 135.22 | 0.545 | 6.42 | -0.907 | 0.905 | -- | -- | -- | 0.228 | 21.7 | -- | -- | -- | -- |
| 147.92 | 0.566 | 6.45 | -1.22 | -- | 0.95 | -- | -- | -0.0288 | -- | 21.4 | 776 | -- | -- |
| 147.77 | 0.566 | 6.45 | -- | 0.851 | 0.471 | -- | -- | 0.162 | 21.4 | 22 | 764 | -- | -- |
| 183.86 | 0.634 | 6.4 | -0.899 | -- | -- | -- | -- | 0.227 | -- | -- | -- | -- | -- |
| 156.91 | 0.586 | 6.4 | -- | 0.903 | -- | -- | -- | 0.329 | 21.6 | -- | -- | -- | -- |
| 184.51 | 0.633 | 6.43 | -- | -- | 0.671 | -- | -- | 0.114 | -- | 22.1 | 765 | -- | -- |
| 205.19 | 0.669 | 6.38 | -- | -- | -- | -- | -- | 0.327 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance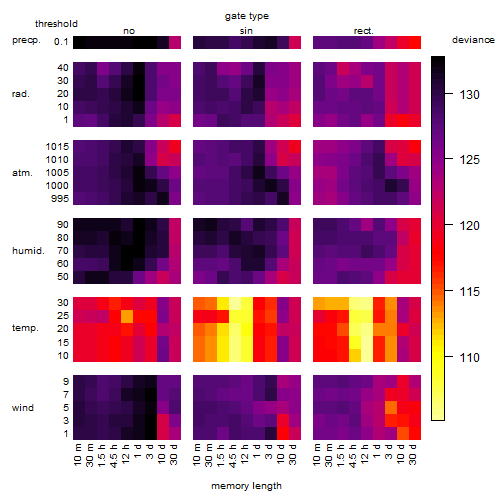
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 105.10 | temperature | 10 | 720 | > th | dose dependent | rect. | 23 | 10 |
| 3 | 105.30 | temperature | 30 | 270 | < th | dose dependent | sin | 6 | NA |
| 4 | 105.35 | temperature | 10 | 720 | > th | dose dependent | rect. | 23 | 12 |
| 10 | 105.74 | temperature | 30 | 720 | < th | dose dependent | rect. | 1 | 7 |
| 13 | 105.90 | temperature | 30 | 720 | < th | dose dependent | rect. | 23 | 9 |
| 16 | 106.03 | temperature | 15 | 270 | > th | dose dependent | sin | 6 | NA |
| 30 | 106.31 | temperature | 30 | 270 | < th | dose dependent | rect. | 2 | 9 |
| 60 | 107.05 | temperature | 15 | 720 | > th | dose dependent | rect. | 23 | 6 |
| 229 | 109.04 | temperature | 25 | 720 | < th | dose dependent | rect. | 3 | 2 |
| 335 | 110.14 | temperature | 20 | 720 | < th | dose independent | rect. | 3 | 3 |
| 432 | 110.95 | temperature | 25 | 720 | < th | dose dependent | rect. | 5 | 1 |
| 461 | 111.14 | temperature | 20 | 720 | > th | dose independent | rect. | 24 | 9 |
| 490 | 111.33 | temperature | 20 | 720 | > th | dose independent | rect. | 23 | 12 |
| 500 | 111.38 | temperature | 20 | 720 | < th | dose independent | rect. | 5 | 1 |
| 662 | 112.33 | temperature | 25 | 270 | < th | dose independent | rect. | 2 | 10 |
| 690 | 112.54 | temperature | 20 | 720 | > th | dose independent | sin | 6 | NA |
| 798 | 113.10 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |
| 814 | 113.20 | temperature | 20 | 720 | > th | dose independent | rect. | 23 | 19 |
| 826 | 113.24 | temperature | 25 | 720 | < th | dose dependent | rect. | 9 | 23 |
| 841 | 113.30 | temperature | 25 | 720 | < th | dose dependent | rect. | 24 | 23 |
| 852 | 113.38 | temperature | 30 | 4320 | < th | dose dependent | rect. | 1 | 1 |
| 854 | 113.39 | temperature | 10 | 4320 | > th | dose dependent | rect. | 1 | 1 |
| 897 | 113.57 | temperature | 20 | 720 | < th | dose independent | sin | 7 | NA |
| 929 | 113.70 | temperature | 25 | 720 | < th | dose dependent | rect. | 3 | 23 |
| 947 | 113.77 | temperature | 25 | 270 | < th | dose independent | sin | 5 | NA |