Os11g0250100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.
FiT-DB / Search/ Help/ Sample detail
|
Os11g0250100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
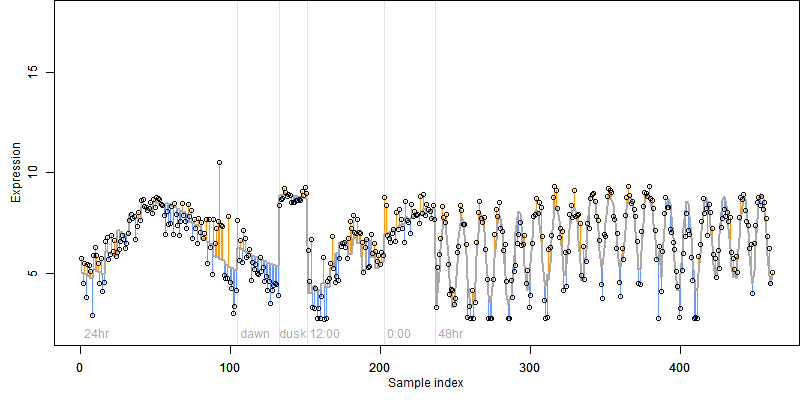
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

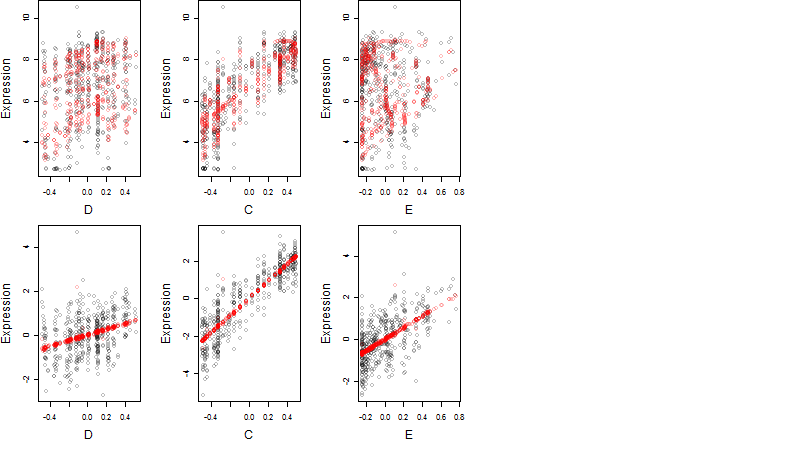
Dependence on each variable
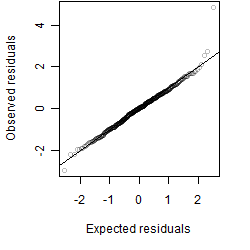
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 352.11 | 0.882 | 6.68 | 1.53 | 4.44 | 2.64 | -1.13 | 1.86 | 0.177 | 20.8 | 21.7 | 333 | 23.4 | -- |
| 354.37 | 0.877 | 6.7 | 1.34 | 4.45 | 2.73 | -1.41 | -- | 0.117 | 20.8 | 21.9 | 332 | 23.5 | -- |
| 355.68 | 0.878 | 6.67 | 1.68 | 4.5 | 2.63 | -- | 2.62 | 0.196 | 20.8 | 21.38 | 325 | 23.3 | -- |
| 360.05 | 0.884 | 6.72 | 1.3 | 4.5 | 2.77 | -- | -- | 0.0815 | 20.7 | 21.7 | 337 | 23.6 | -- |
| 550.94 | 1.09 | 6.38 | 0.922 | -- | 4.46 | -- | 4.57 | 0.748 | -- | -15.1 | 961 | 18.3 | -- |
| 459.82 | 1.01 | 6.6 | 1.73 | 3.89 | -- | -1.61 | -- | 0.515 | 20 | -- | -- | -- | -- |
| 467.58 | 1.01 | 6.6 | 1.78 | 3.89 | -- | -- | -- | 0.514 | 20 | -- | -- | -- | -- |
| 448.63 | 0.986 | 6.53 | 1.44 | -- | 4.58 | -- | -- | 0.37 | -- | -28.08 | 886 | 17.5 | -- |
| 401.84 | 0.934 | 6.77 | -- | 4.63 | 3.15 | -- | -- | -0.128 | 20.8 | 21.5 | 351 | 23.7 | -- |
| 1295.22 | 1.68 | 6.5 | 1.74 | -- | -- | -- | -- | 0.465 | -- | -- | -- | -- | -- |
| 550.72 | 1.1 | 6.65 | -- | 3.88 | -- | -- | -- | 0.316 | 20 | -- | -- | -- | -- |
| 486.8 | 1.03 | 6.58 | -- | -- | 5.09 | -- | -- | 0.128 | -- | -92.18 | 1260 | 20.7 | -- |
| 1374.93 | 1.73 | 6.55 | -- | -- | -- | -- | -- | 0.272 | -- | -- | -- | -- | -- |
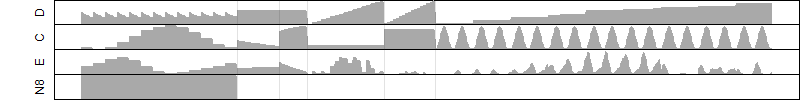
Results of the grid search
Summarized heatmap of deviance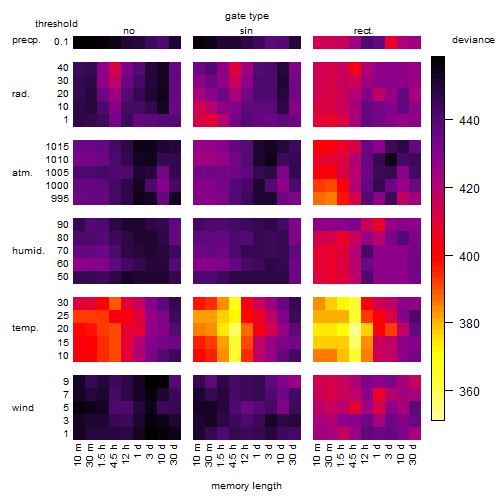
|
Histogram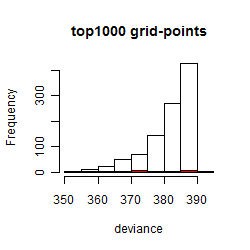 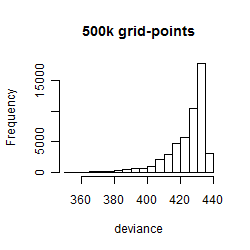
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 351.07 | temperature | 20 | 270 | > th | dose dependent | rect. | 23 | 16 |
| 8 | 356.33 | temperature | 20 | 270 | > th | dose dependent | sin | 4 | NA |
| 25 | 363.20 | temperature | 30 | 270 | < th | dose dependent | rect. | 1 | 21 |
| 30 | 364.14 | temperature | 25 | 270 | > th | dose independent | rect. | 23 | 17 |
| 56 | 367.68 | temperature | 30 | 270 | < th | dose dependent | sin | 5 | NA |
| 66 | 368.19 | temperature | 20 | 270 | > th | dose dependent | rect. | 7 | 9 |
| 74 | 369.02 | temperature | 25 | 270 | < th | dose independent | rect. | 3 | 9 |
| 85 | 369.96 | temperature | 10 | 270 | > th | dose dependent | rect. | 6 | 22 |
| 105 | 372.28 | temperature | 30 | 90 | < th | dose dependent | rect. | 5 | 18 |
| 116 | 373.11 | temperature | 30 | 270 | < th | dose dependent | rect. | 3 | 8 |
| 121 | 373.27 | temperature | 30 | 270 | < th | dose dependent | rect. | 2 | 11 |
| 124 | 373.33 | temperature | 20 | 30 | > th | dose dependent | rect. | 8 | 21 |
| 151 | 374.71 | temperature | 25 | 270 | < th | dose independent | sin | 4 | NA |
| 165 | 375.46 | temperature | 25 | 270 | > th | dose independent | sin | 3 | NA |
| 209 | 377.08 | temperature | 30 | 90 | < th | dose independent | rect. | 5 | 18 |
| 210 | 377.12 | temperature | 25 | 270 | > th | dose independent | rect. | 7 | 9 |
| 218 | 377.40 | temperature | 25 | 270 | < th | dose independent | rect. | 3 | 17 |
| 318 | 380.34 | temperature | 25 | 10 | > th | dose dependent | rect. | 8 | 12 |
| 394 | 382.15 | temperature | 30 | 90 | < th | dose dependent | rect. | 5 | 8 |
| 521 | 384.37 | temperature | 25 | 10 | > th | dose dependent | rect. | 8 | 16 |
| 662 | 386.20 | atmosphere | 995 | 30 | > th | dose dependent | rect. | 5 | 18 |
| 730 | 387.06 | temperature | 20 | 720 | > th | dose dependent | rect. | 8 | 1 |
| 925 | 389.38 | temperature | 30 | 270 | < th | dose dependent | rect. | 14 | 23 |
| 938 | 389.52 | temperature | 30 | 270 | < th | dose dependent | rect. | 16 | 23 |
| 939 | 389.54 | temperature | 30 | 270 | < th | dose dependent | no | NA | NA |
| 967 | 389.82 | temperature | 10 | 270 | > th | dose dependent | no | NA | NA |