Os11g0242200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein phosphatase 2C family protein.
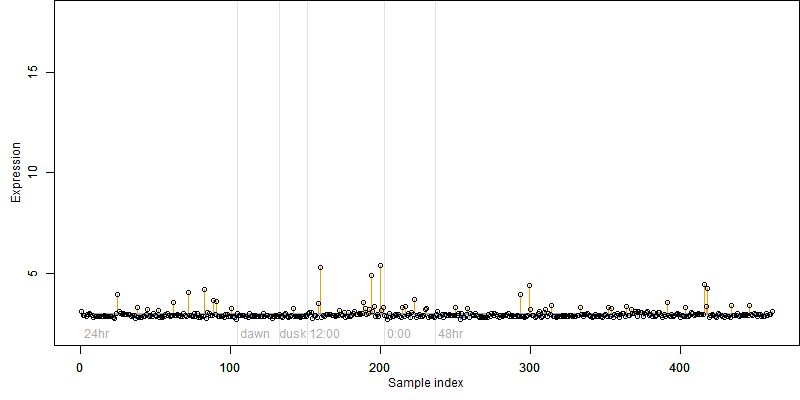

FiT-DB / Search/ Help/ Sample detail
|
Os11g0242200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein phosphatase 2C family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
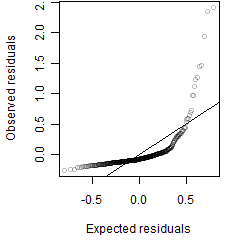
Dependence on each variable

Residual plot

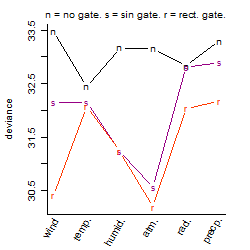
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 31.94 | 0.266 | 2.97 | 0.131 | 0.0164 | 1.14 | 0.254 | 2.28 | 0.0102 | 18.7 | 19 | 600 | -- | -- |
| 34 | 0.272 | 2.95 | 0.211 | 0.0409 | 0.285 | 0.234 | -- | 0.029 | 16.9 | 21.81 | 785 | -- | -- |
| 31.86 | 0.263 | 2.97 | 0.126 | 0.0213 | 1.88 | -- | 3.72 | -9.61e-05 | 19 | 18.08 | 477 | -- | -- |
| 34.08 | 0.272 | 2.94 | 0.172 | 0.0543 | 0.34 | -- | -- | 0.0328 | 15.3 | 20.3 | 691 | -- | -- |
| 31.82 | 0.263 | 2.97 | 0.129 | -- | 2.03 | -- | 4.17 | 0.00397 | -- | 17.96 | 476 | -- | -- |
| 34.51 | 0.275 | 2.95 | 0.113 | 0.0804 | -- | 0.169 | -- | -0.0037 | 13.6 | -- | -- | -- | -- |
| 34.61 | 0.275 | 2.95 | 0.119 | 0.0807 | -- | -- | -- | -0.00297 | 12.9 | -- | -- | -- | -- |
| 34.24 | 0.273 | 2.94 | 0.182 | -- | 0.314 | -- | -- | 0.0237 | -- | 20.2 | 713 | -- | -- |
| 34.72 | 0.274 | 2.95 | -- | 0.0689 | 0.186 | -- | -- | -0.00669 | 13.4 | 19.6 | 621 | -- | -- |
| 35.01 | 0.276 | 2.95 | 0.116 | -- | -- | -- | -- | -0.00451 | -- | -- | -- | -- | -- |
| 34.98 | 0.277 | 2.95 | -- | 0.0788 | -- | -- | -- | -0.0162 | 12.9 | -- | -- | -- | -- |
| 35 | 0.276 | 2.95 | -- | -- | 0.208 | -- | -- | -0.00522 | -- | 19.86 | 688 | -- | -- |
| 35.37 | 0.278 | 2.95 | -- | -- | -- | -- | -- | -0.0173 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance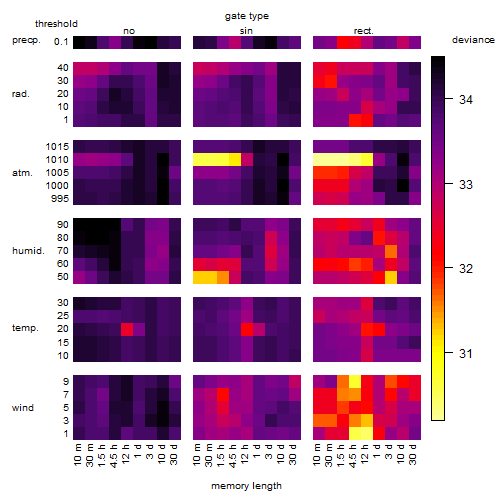
|
Histogram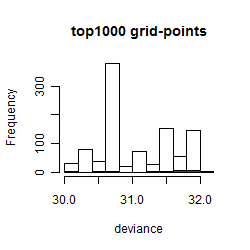 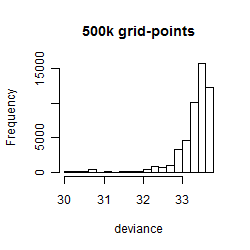
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 30.19 | atmosphere | 1010 | 10 | > th | dose dependent | rect. | 12 | 1 |
| 111 | 30.41 | wind | 1 | 720 | < th | dose dependent | rect. | 8 | 2 |
| 125 | 30.53 | wind | 1 | 720 | < th | dose independent | rect. | 8 | 2 |
| 127 | 30.56 | atmosphere | 1010 | 30 | > th | dose dependent | sin | 23 | NA |
| 382 | 30.70 | wind | 9 | 270 | > th | dose independent | rect. | 9 | 1 |
| 617 | 31.23 | humidity | 50 | 10 | > th | dose independent | sin | 8 | NA |
| 618 | 31.24 | humidity | 50 | 4320 | > th | dose independent | rect. | 16 | 1 |
| 627 | 31.26 | wind | 1 | 270 | < th | dose independent | rect. | 17 | 1 |
| 629 | 31.26 | wind | 1 | 270 | < th | dose dependent | rect. | 17 | 1 |
| 654 | 31.45 | atmosphere | 1010 | 30 | > th | dose independent | rect. | 11 | 1 |
| 778 | 31.52 | wind | 7 | 270 | > th | dose dependent | rect. | 9 | 1 |
| 783 | 31.54 | wind | 7 | 90 | > th | dose independent | rect. | 20 | 15 |
| 798 | 31.62 | humidity | 70 | 4320 | > th | dose independent | rect. | 21 | 7 |
| 801 | 31.63 | wind | 9 | 90 | > th | dose dependent | rect. | 20 | 14 |
| 802 | 31.64 | humidity | 70 | 4320 | > th | dose independent | rect. | 21 | 2 |
| 833 | 31.70 | wind | 3 | 10 | < th | dose dependent | rect. | 12 | 1 |
| 850 | 31.79 | wind | 9 | 4320 | > th | dose dependent | rect. | 14 | 1 |
| 861 | 31.83 | humidity | 60 | 270 | < th | dose dependent | rect. | 10 | 1 |
| 891 | 31.89 | humidity | 60 | 270 | < th | dose independent | rect. | 8 | 3 |
| 910 | 31.92 | humidity | 70 | 4320 | > th | dose independent | rect. | 19 | 12 |
| 946 | 31.99 | humidity | 70 | 4320 | < th | dose independent | rect. | 20 | 1 |
| 957 | 31.99 | humidity | 70 | 4320 | < th | dose dependent | rect. | 20 | 1 |
| 997 | 32.01 | atmosphere | 1010 | 30 | > th | dose independent | sin | 21 | NA |