Os11g0229500
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to NBS-LRR protein (Fragment).


FiT-DB / Search/ Help/ Sample detail
|
Os11g0229500 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to NBS-LRR protein (Fragment).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

Dependence on each variable
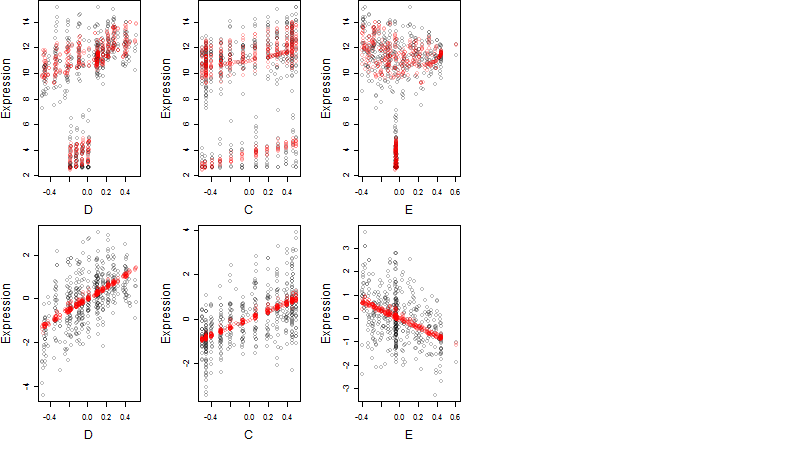
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 441.24 | 0.987 | 11.4 | 2.68 | 1.66 | -1.83 | -0.857 | 0.824 | -7.68 | 1.63 | 81.63 | 1419 | -- | -- |
| 441.62 | 0.979 | 11.5 | 2.63 | 1.77 | -1.82 | -0.799 | -- | -7.68 | 1.61 | 82.09 | 1336 | -- | -- |
| 444.1 | 0.982 | 11.4 | 2.71 | 1.66 | -1.85 | -- | 0.78 | -7.67 | 1.58 | 81.95 | 1420 | -- | -- |
| 444.39 | 0.982 | 11.5 | 2.64 | 1.78 | -1.84 | -- | -- | -7.67 | 1.56 | 82.55 | 1336 | -- | -- |
| 526.35 | 1.07 | 11.4 | 2.4 | -- | -2.46 | -- | 1.17 | -7.56 | -- | 136.1 | 8 | -- | -- |
| 519.11 | 1.07 | 11.4 | 2.31 | 1.58 | -- | -1.18 | -- | -7.63 | 1.57 | -- | -- | -- | -- |
| 524.61 | 1.07 | 11.4 | 2.35 | 1.58 | -- | -- | -- | -7.62 | 1.48 | -- | -- | -- | -- |
| 528.7 | 1.08 | 11.4 | 2.39 | -- | -2.41 | -- | -- | -7.57 | -- | 136.1 | 8 | -- | -- |
| 615.53 | 1.16 | 11.5 | -- | 1.86 | -1.63 | -- | -- | -7.84 | 1.58 | 133.5 | 1274 | -- | -- |
| 676.79 | 1.22 | 11.4 | 2.42 | -- | -- | -- | -- | -7.59 | -- | -- | -- | -- | -- |
| 670.58 | 1.21 | 11.5 | -- | 1.62 | -- | -- | -- | -7.88 | 1.53 | -- | -- | -- | -- |
| 677.81 | 1.21 | 11.5 | -- | -- | -2.41 | -- | -- | -7.83 | -- | 183.3 | 2 | -- | -- |
| 830.78 | 1.35 | 11.5 | -- | -- | -- | -- | -- | -7.85 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 439.97 | humidity | 80 | 1440 | < th | dose dependent | rect. | 12 | 22 |
| 2 | 440.10 | humidity | 60 | 1440 | > th | dose independent | rect. | 15 | 2 |
| 7 | 440.64 | humidity | 80 | 1440 | < th | dose dependent | rect. | 19 | 22 |
| 14 | 440.95 | humidity | 90 | 1440 | < th | dose dependent | rect. | 13 | 1 |
| 20 | 441.29 | humidity | 80 | 1440 | < th | dose dependent | rect. | 12 | 5 |
| 22 | 441.45 | humidity | 80 | 1440 | < th | dose dependent | rect. | 6 | 20 |
| 24 | 441.45 | humidity | 80 | 1440 | < th | dose dependent | rect. | 24 | 22 |
| 25 | 441.46 | humidity | 80 | 1440 | < th | dose dependent | rect. | 6 | 16 |
| 50 | 441.52 | humidity | 80 | 1440 | < th | dose dependent | no | NA | NA |
| 60 | 441.55 | humidity | 80 | 1440 | < th | dose dependent | rect. | 11 | 15 |
| 69 | 441.60 | humidity | 80 | 1440 | < th | dose dependent | sin | 21 | NA |
| 71 | 441.60 | humidity | 80 | 1440 | < th | dose dependent | rect. | 11 | 11 |
| 89 | 441.76 | humidity | 60 | 1440 | < th | dose independent | rect. | 14 | 3 |
| 121 | 442.11 | humidity | 80 | 1440 | < th | dose dependent | rect. | 23 | 19 |
| 129 | 442.17 | humidity | 80 | 1440 | < th | dose dependent | rect. | 6 | 12 |
| 158 | 442.34 | humidity | 80 | 1440 | < th | dose dependent | rect. | 15 | 23 |
| 213 | 442.82 | humidity | 50 | 1440 | > th | dose dependent | rect. | 13 | 1 |
| 226 | 443.04 | humidity | 60 | 1440 | > th | dose independent | rect. | 14 | 20 |
| 280 | 444.18 | humidity | 50 | 1440 | > th | dose dependent | rect. | 12 | 5 |
| 324 | 444.73 | humidity | 60 | 1440 | < th | dose independent | rect. | 14 | 20 |
| 371 | 445.41 | humidity | 60 | 1440 | > th | dose independent | rect. | 14 | 7 |
| 375 | 445.45 | humidity | 60 | 1440 | > th | dose independent | rect. | 14 | 9 |
| 391 | 445.54 | humidity | 60 | 1440 | > th | dose independent | rect. | 14 | 15 |
| 410 | 445.82 | humidity | 50 | 1440 | > th | dose dependent | rect. | 11 | 11 |
| 754 | 447.45 | humidity | 50 | 720 | > th | dose dependent | rect. | 15 | 9 |
| 776 | 447.64 | humidity | 50 | 1440 | > th | dose dependent | sin | 20 | NA |
| 914 | 448.61 | humidity | 50 | 1440 | > th | dose dependent | rect. | 15 | 1 |
| 920 | 448.67 | humidity | 70 | 1440 | > th | dose independent | rect. | 17 | 22 |
| 972 | 449.33 | humidity | 70 | 1440 | < th | dose independent | rect. | 17 | 22 |