Os11g0181800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Short-chain dehydrogenase Tic32.

FiT-DB / Search/ Help/ Sample detail
|
Os11g0181800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Short-chain dehydrogenase Tic32.
|
|
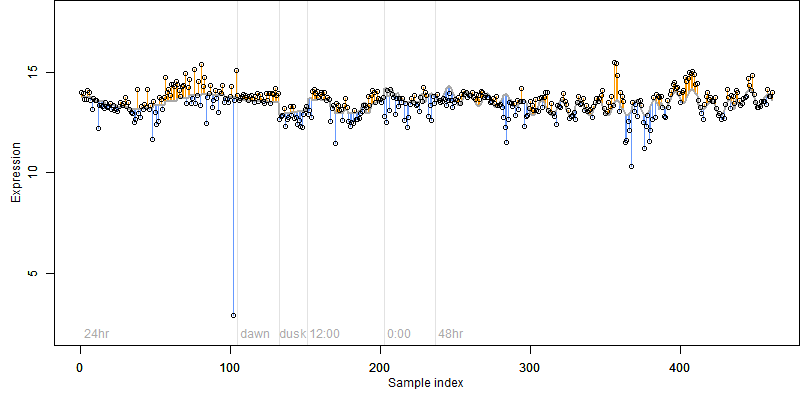
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

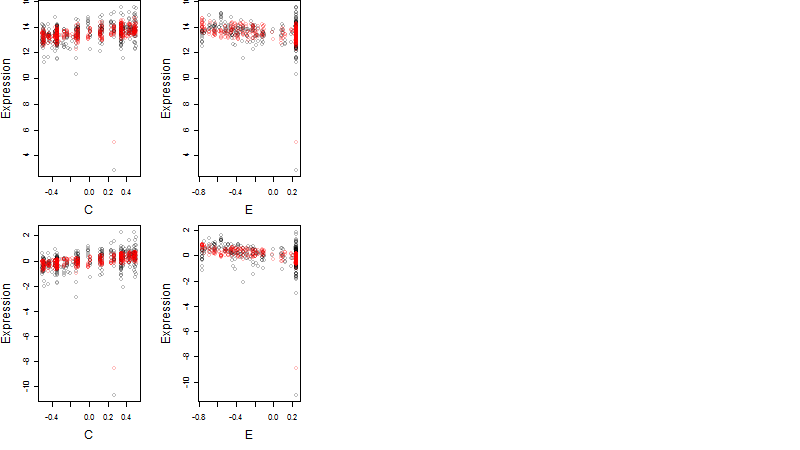
Dependence on each variable
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 243.97 | 0.734 | 13.4 | 0.191 | 0.669 | -1.1 | 1.58 | -1.53 | 0.261 | 3.91 | 20.7 | 1314 | -- | -- |
| 245.32 | 0.729 | 13.4 | 0.378 | 0.698 | -0.749 | 1.59 | -- | 0.375 | 3.85 | 22.6 | 1243 | -- | -- |
| 250.57 | 0.737 | 13.4 | 0.217 | 0.678 | -1.1 | -- | -1.31 | 0.269 | 3.36 | 20.79 | 1303 | -- | -- |
| 251.22 | 0.738 | 13.4 | 0.354 | 0.709 | -0.777 | -- | -- | 0.365 | 3.13 | 22.4 | 1134 | -- | -- |
| 274.39 | 0.771 | 13.5 | 0.238 | -- | -1.09 | -- | -1.42 | 0.277 | -- | 20.7 | 1761 | -- | -- |
| 269.11 | 0.769 | 13.5 | 0.111 | 0.62 | -- | 1.69 | -- | 0.104 | 4.21 | -- | -- | -- | -- |
| 276.32 | 0.778 | 13.5 | 0.0924 | 0.631 | -- | -- | -- | 0.0952 | 3.55 | -- | -- | -- | -- |
| 278.09 | 0.777 | 13.4 | 0.418 | -- | -0.751 | -- | -- | 0.364 | -- | 22.13 | 1744 | -- | -- |
| 254.36 | 0.743 | 13.4 | -- | 0.733 | -0.722 | -- | -- | 0.315 | 3.13 | 22.4 | 1121 | -- | -- |
| 298.53 | 0.807 | 13.5 | 0.119 | -- | -- | -- | -- | 0.112 | -- | -- | -- | -- | -- |
| 276.54 | 0.778 | 13.5 | -- | 0.633 | -- | -- | -- | 0.0849 | 3.54 | -- | -- | -- | -- |
| 281.88 | 0.782 | 13.4 | -- | -- | -0.635 | -- | -- | 0.293 | -- | 22.34 | 1500 | -- | -- |
| 298.91 | 0.807 | 13.5 | -- | -- | -- | -- | -- | 0.0988 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance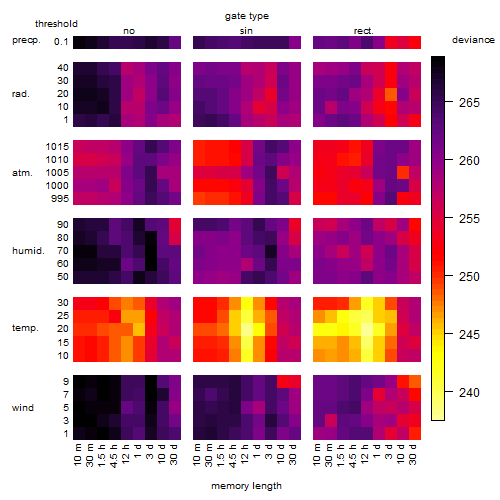
|
Histogram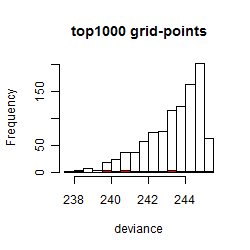 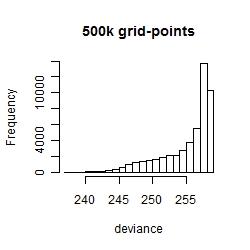
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 237.51 | temperature | 20 | 720 | > th | dose dependent | rect. | 24 | 14 |
| 4 | 238.05 | temperature | 20 | 720 | > th | dose dependent | sin | 6 | NA |
| 19 | 239.63 | temperature | 15 | 720 | > th | dose dependent | rect. | 21 | 18 |
| 20 | 239.64 | temperature | 25 | 720 | > th | dose independent | rect. | 24 | 11 |
| 28 | 239.90 | temperature | 25 | 720 | > th | dose independent | rect. | 24 | 13 |
| 30 | 239.91 | temperature | 20 | 720 | > th | dose dependent | rect. | 1 | 8 |
| 52 | 240.33 | temperature | 25 | 720 | < th | dose dependent | rect. | 4 | 15 |
| 60 | 240.51 | temperature | 25 | 720 | > th | dose independent | rect. | 1 | 8 |
| 70 | 240.67 | temperature | 25 | 270 | > th | dose independent | rect. | 3 | 16 |
| 77 | 240.81 | temperature | 25 | 720 | < th | dose dependent | sin | 1 | NA |
| 129 | 241.48 | temperature | 25 | 720 | > th | dose independent | sin | 6 | NA |
| 147 | 241.66 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 20 |
| 177 | 241.92 | temperature | 20 | 1440 | > th | dose independent | rect. | 20 | 10 |
| 199 | 242.11 | temperature | 20 | 1440 | < th | dose independent | rect. | 20 | 10 |
| 280 | 242.63 | temperature | 30 | 720 | < th | dose dependent | sin | 4 | NA |
| 295 | 242.72 | temperature | 20 | 30 | > th | dose dependent | rect. | 3 | 17 |
| 340 | 243.03 | temperature | 20 | 1440 | < th | dose independent | rect. | 24 | 6 |
| 344 | 243.04 | temperature | 25 | 720 | < th | dose independent | rect. | 24 | 13 |
| 421 | 243.39 | temperature | 20 | 1440 | > th | dose independent | sin | 12 | NA |
| 531 | 243.86 | temperature | 20 | 1440 | < th | dose independent | sin | 12 | NA |
| 567 | 243.98 | temperature | 25 | 720 | < th | dose independent | sin | 5 | NA |
| 599 | 244.08 | temperature | 20 | 1440 | > th | dose dependent | rect. | 24 | 4 |
| 634 | 244.18 | temperature | 20 | 1440 | < th | dose independent | rect. | 3 | 1 |
| 929 | 244.99 | temperature | 20 | 1440 | > th | dose independent | rect. | 5 | 1 |
| 972 | 245.08 | temperature | 25 | 270 | > th | dose independent | rect. | 8 | 11 |
| 978 | 245.10 | temperature | 25 | 270 | > th | dose independent | sin | 23 | NA |