Os11g0111200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.
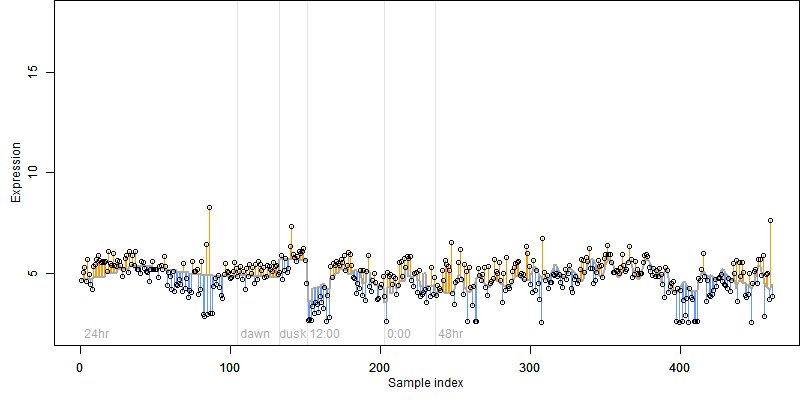

FiT-DB / Search/ Help/ Sample detail
|
Os11g0111200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

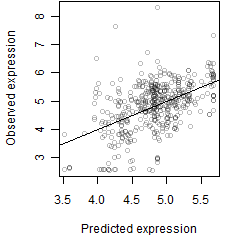
Dependence on each variable

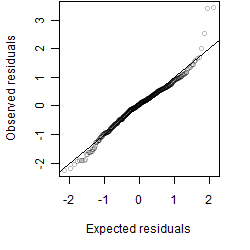
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 240.41 | 0.728 | 4.84 | -0.37 | 0.541 | -2.53 | 1.34 | -0.576 | -0.149 | 11.3 | 31.57 | 718 | -- | -- |
| 240.67 | 0.723 | 4.85 | -0.467 | 0.543 | -2.52 | 1.26 | -- | -0.162 | 11.5 | 31.95 | 729 | -- | -- |
| 246.92 | 0.732 | 4.86 | -0.512 | 0.526 | -2.59 | -- | 0.514 | -0.201 | 10.7 | 31.7 | 676 | -- | -- |
| 246.84 | 0.732 | 4.85 | -0.413 | 0.498 | -2.59 | -- | -- | -0.183 | 9.87 | 31.53 | 611 | -- | -- |
| 255.89 | 0.745 | 4.84 | -0.253 | -- | -2.23 | -- | -0.208 | -0.114 | -- | 31.2 | 412 | -- | -- |
| 326.23 | 0.847 | 4.77 | 0.211 | 0.319 | -- | 1.32 | -- | 0.258 | 15 | -- | -- | -- | -- |
| 331.05 | 0.852 | 4.77 | 0.24 | 0.362 | -- | -- | -- | 0.267 | 16.4 | -- | -- | -- | -- |
| 255.93 | 0.745 | 4.85 | -0.289 | -- | -2.24 | -- | -- | -0.126 | -- | 31.3 | 412 | -- | -- |
| 250.76 | 0.738 | 4.83 | -- | 0.446 | -2.42 | -- | -- | -0.108 | 9.81 | 31.79 | 585 | -- | -- |
| 338.09 | 0.859 | 4.76 | 0.226 | -- | -- | -- | -- | 0.257 | -- | -- | -- | -- | -- |
| 332.57 | 0.853 | 4.77 | -- | 0.358 | -- | -- | -- | 0.24 | 16.5 | -- | -- | -- | -- |
| 257.95 | 0.748 | 4.84 | -- | -- | -2.15 | -- | -- | -0.0746 | -- | 31.51 | 400 | -- | -- |
| 339.44 | 0.86 | 4.77 | -- | -- | -- | -- | -- | 0.232 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance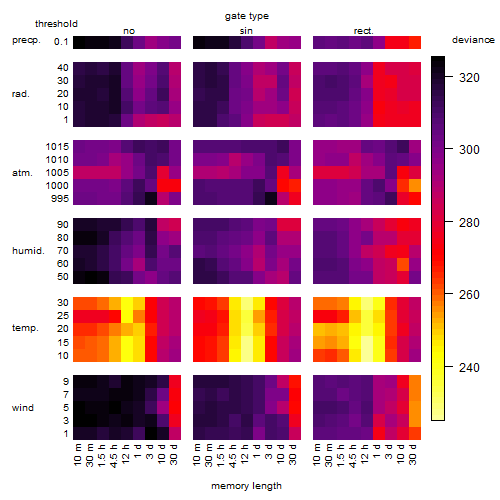
|
Histogram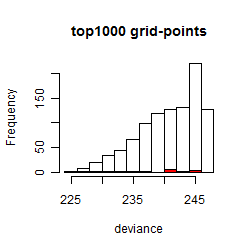 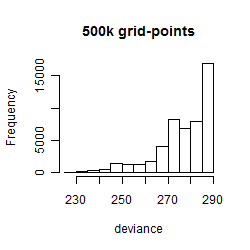
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 225.02 | temperature | 10 | 720 | > th | dose dependent | rect. | 21 | 18 |
| 7 | 227.32 | temperature | 10 | 720 | > th | dose dependent | sin | 6 | NA |
| 21 | 229.10 | temperature | 30 | 720 | < th | dose dependent | sin | 6 | NA |
| 23 | 229.47 | temperature | 30 | 720 | < th | dose dependent | rect. | 23 | 15 |
| 41 | 231.05 | temperature | 30 | 720 | < th | dose dependent | rect. | 24 | 19 |
| 94 | 233.24 | temperature | 30 | 720 | < th | dose dependent | rect. | 1 | 10 |
| 153 | 235.58 | temperature | 10 | 720 | > th | dose dependent | rect. | 3 | 8 |
| 241 | 237.37 | temperature | 10 | 720 | > th | dose dependent | rect. | 12 | 23 |
| 413 | 240.34 | temperature | 10 | 1440 | > th | dose dependent | rect. | 8 | 1 |
| 432 | 240.57 | temperature | 30 | 1440 | < th | dose dependent | rect. | 8 | 1 |
| 459 | 241.09 | temperature | 30 | 720 | < th | dose dependent | no | NA | NA |
| 471 | 241.26 | temperature | 10 | 720 | > th | dose dependent | rect. | 1 | 23 |
| 472 | 241.28 | temperature | 10 | 720 | > th | dose dependent | no | NA | NA |
| 513 | 241.92 | temperature | 10 | 720 | > th | dose dependent | rect. | 3 | 23 |
| 573 | 242.90 | temperature | 30 | 720 | < th | dose dependent | rect. | 7 | 23 |
| 735 | 244.81 | temperature | 15 | 720 | > th | dose dependent | rect. | 7 | 23 |
| 787 | 245.39 | temperature | 10 | 270 | > th | dose dependent | sin | 1 | NA |
| 800 | 245.48 | temperature | 30 | 1440 | < th | dose dependent | rect. | 7 | 15 |
| 868 | 245.98 | temperature | 10 | 1440 | > th | dose dependent | rect. | 5 | 17 |