Os11g0108700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Laccase (Diphenol oxidase).
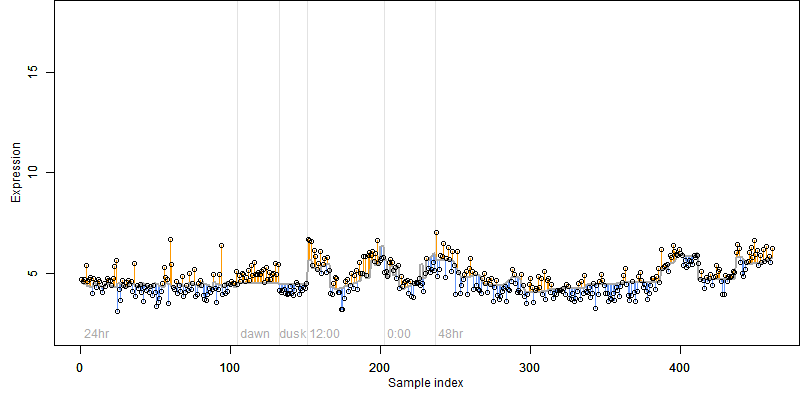

FiT-DB / Search/ Help/ Sample detail
|
Os11g0108700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Laccase (Diphenol oxidase).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
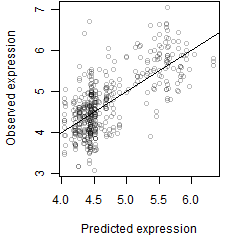
Dependence on each variable

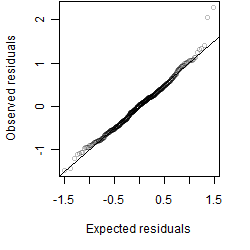
Residual plot
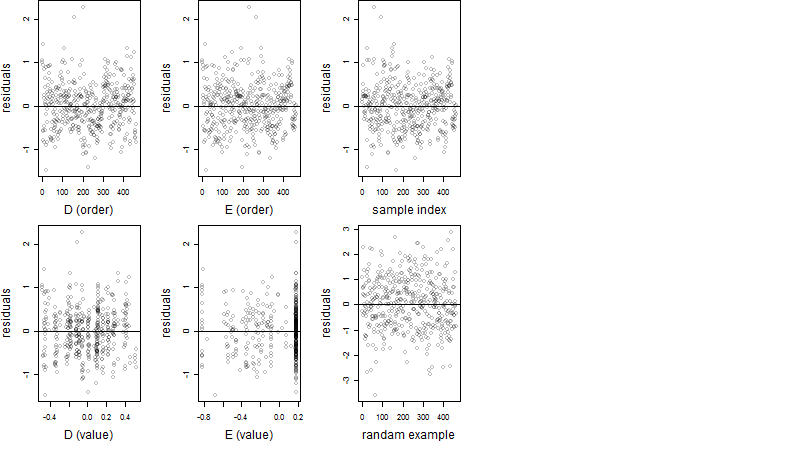
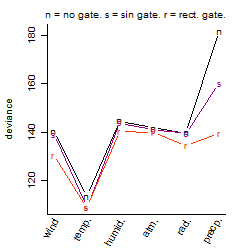
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 108.13 | 0.489 | 4.63 | 1.49 | 0.523 | -1.85 | 0.787 | 0.843 | 0.222 | 7.78 | 21.39 | 1366 | -- | -- |
| 109.9 | 0.488 | 4.66 | 1.3 | 0.539 | -1.86 | 0.748 | -- | 0.172 | 7.76 | 21.42 | 1389 | -- | -- |
| 109.9 | 0.488 | 4.63 | 1.5 | 0.525 | -1.85 | -- | 0.834 | 0.221 | 7.65 | 21.39 | 1366 | -- | -- |
| 111.4 | 0.492 | 4.66 | 1.3 | 0.525 | -1.92 | -- | -- | 0.175 | 7.67 | 21.4 | 1367 | -- | -- |
| 124.51 | 0.52 | 4.64 | 1.49 | -- | -1.81 | -- | 0.88 | 0.224 | -- | 21.3 | 1248 | -- | -- |
| 213.81 | 0.685 | 4.78 | 0.482 | 0.529 | -- | 0.746 | -- | -0.357 | 8.25 | -- | -- | -- | -- |
| 215.51 | 0.687 | 4.78 | 0.505 | 0.526 | -- | -- | -- | -0.357 | 8.37 | -- | -- | -- | -- |
| 126.23 | 0.523 | 4.66 | 1.27 | -- | -1.91 | -- | -- | 0.183 | -- | 21.4 | 1349 | -- | -- |
| 148.97 | 0.568 | 4.71 | -- | 0.519 | -1.46 | -- | -- | -0.0675 | 7.42 | 21.4 | 1275 | -- | -- |
| 230.91 | 0.71 | 4.79 | 0.508 | -- | -- | -- | -- | -0.351 | -- | -- | -- | -- | -- |
| 222.22 | 0.697 | 4.79 | -- | 0.529 | -- | -- | -- | -0.413 | 8.3 | -- | -- | -- | -- |
| 154.78 | 0.579 | 4.72 | -- | -- | -1.34 | -- | -- | -0.0262 | -- | 22.05 | 854 | -- | -- |
| 237.71 | 0.72 | 4.8 | -- | -- | -- | -- | -- | -0.407 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance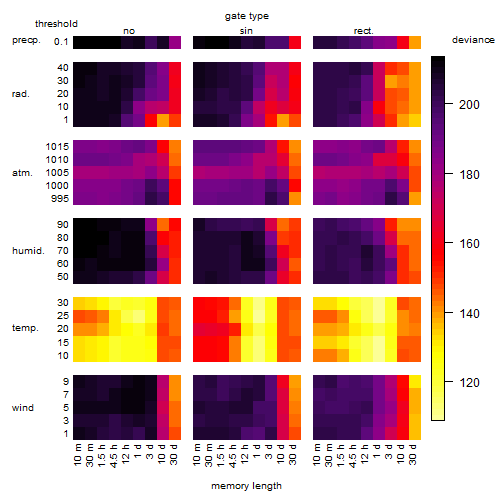
|
Histogram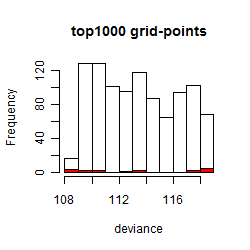 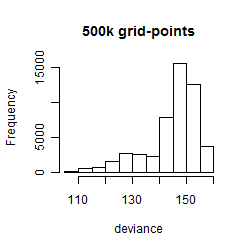
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 108.77 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 9 |
| 2 | 108.78 | temperature | 25 | 1440 | < th | dose dependent | sin | 9 | NA |
| 5 | 108.84 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 14 |
| 24 | 109.10 | temperature | 30 | 1440 | < th | dose dependent | rect. | 1 | 5 |
| 25 | 109.11 | temperature | 10 | 1440 | > th | dose dependent | rect. | 1 | 5 |
| 151 | 110.09 | temperature | 30 | 1440 | < th | dose dependent | rect. | 5 | 1 |
| 205 | 110.49 | temperature | 15 | 1440 | > th | dose dependent | rect. | 5 | 1 |
| 411 | 112.39 | temperature | 15 | 1440 | > th | dose dependent | sin | 9 | NA |
| 519 | 113.38 | temperature | 25 | 1440 | < th | dose dependent | no | NA | NA |
| 558 | 113.75 | temperature | 25 | 1440 | < th | dose dependent | rect. | 24 | 23 |
| 852 | 117.27 | temperature | 20 | 1440 | > th | dose independent | rect. | 20 | 16 |
| 906 | 117.77 | temperature | 20 | 1440 | < th | dose independent | rect. | 20 | 16 |
| 941 | 118.05 | temperature | 20 | 1440 | > th | dose independent | sin | 7 | NA |
| 951 | 118.10 | temperature | 20 | 1440 | < th | dose independent | rect. | 1 | 10 |
| 968 | 118.17 | temperature | 20 | 1440 | > th | dose independent | rect. | 1 | 11 |
| 984 | 118.27 | temperature | 20 | 1440 | < th | dose independent | sin | 7 | NA |