Os10g0537500
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF1084 family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os10g0537500 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF1084 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

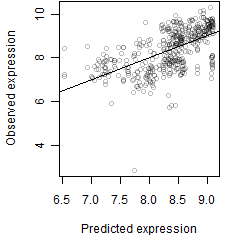
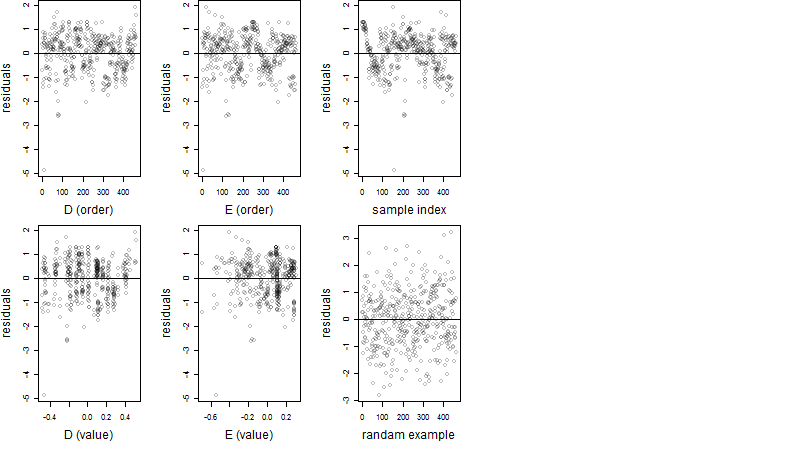
Dependence on each variable

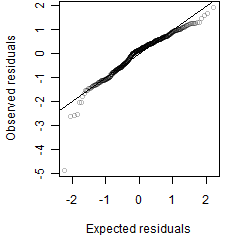
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 244.67 | 0.735 | 8.54 | -2.29 | 1.33 | 3.15 | -1.73 | -4.18 | -0.728 | 7 | 16.04 | 646 | -- | -- |
| 255.85 | 0.745 | 8.44 | -1.53 | 1.33 | 3.24 | -0.814 | -- | -0.517 | 6.86 | 14.8 | 613 | -- | -- |
| 251.07 | 0.738 | 8.51 | -2.17 | 1.36 | 3.18 | -- | -3.2 | -0.669 | 7.03 | 16.17 | 637 | -- | -- |
| 257.27 | 0.747 | 8.44 | -1.56 | 1.38 | 3.35 | -- | -- | -0.524 | 6.89 | 14.41 | 609 | -- | -- |
| 266.05 | 0.76 | 8.56 | -2.22 | -- | 2.68 | -- | -3.05 | -0.657 | -- | 15.34 | 1354 | -- | -- |
| 385.4 | 0.92 | 8.36 | -0.745 | 0.561 | -- | -1.56 | -- | -0.0226 | 5.7 | -- | -- | -- | -- |
| 391.31 | 0.926 | 8.35 | -0.753 | 0.552 | -- | -- | -- | -0.0159 | 5.88 | -- | -- | -- | -- |
| 278.11 | 0.777 | 8.47 | -1.6 | -- | 2.73 | -- | -- | -0.506 | -- | 10.1 | 1374 | -- | -- |
| 313.63 | 0.825 | 8.39 | -- | 1.25 | 2.36 | -- | -- | -0.232 | 6.42 | 17.14 | 531 | -- | -- |
| 407.13 | 0.943 | 8.36 | -0.737 | -- | -- | -- | -- | -0.00304 | -- | -- | -- | -- | -- |
| 406.26 | 0.943 | 8.33 | -- | 0.541 | -- | -- | -- | 0.068 | 5.95 | -- | -- | -- | -- |
| 336.76 | 0.855 | 8.41 | -- | -- | 2.07 | -- | -- | -0.228 | -- | 12.32 | 1338 | -- | -- |
| 421.44 | 0.958 | 8.34 | -- | -- | -- | -- | -- | 0.0787 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance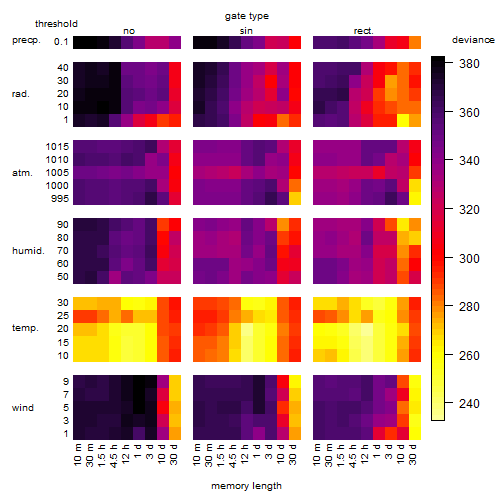
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 232.67 | temperature | 20 | 720 | > th | dose dependent | rect. | 21 | 19 |
| 2 | 233.06 | temperature | 20 | 720 | > th | dose dependent | rect. | 23 | 17 |
| 17 | 236.73 | temperature | 20 | 720 | > th | dose dependent | sin | 4 | NA |
| 46 | 239.99 | temperature | 20 | 270 | > th | dose dependent | rect. | 1 | 21 |
| 47 | 240.08 | temperature | 20 | 270 | > th | dose dependent | rect. | 3 | 19 |
| 149 | 244.56 | temperature | 20 | 270 | > th | dose dependent | rect. | 12 | 23 |
| 225 | 246.80 | temperature | 30 | 1440 | < th | dose dependent | rect. | 18 | 1 |
| 231 | 247.00 | temperature | 15 | 720 | > th | dose dependent | no | NA | NA |
| 239 | 247.19 | temperature | 20 | 1440 | > th | dose dependent | rect. | 18 | 1 |
| 246 | 247.33 | temperature | 10 | 1440 | > th | dose dependent | rect. | 18 | 1 |
| 272 | 247.90 | temperature | 15 | 720 | > th | dose dependent | rect. | 14 | 23 |
| 298 | 248.66 | temperature | 20 | 1440 | > th | dose dependent | rect. | 13 | 20 |
| 322 | 249.06 | temperature | 10 | 720 | > th | dose dependent | rect. | 10 | 23 |
| 397 | 249.95 | temperature | 20 | 1440 | > th | dose dependent | sin | 16 | NA |
| 435 | 250.17 | temperature | 20 | 1440 | > th | dose dependent | rect. | 8 | 23 |
| 470 | 250.36 | temperature | 20 | 1440 | > th | dose dependent | rect. | 14 | 14 |
| 492 | 250.44 | temperature | 20 | 1440 | > th | dose dependent | rect. | 15 | 11 |