Os10g0469600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Leucine rich repeat, N-terminal domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os10g0469600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Leucine rich repeat, N-terminal domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
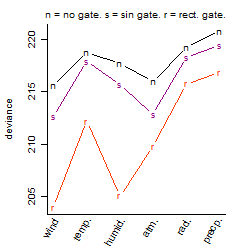
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 215.96 | 0.69 | 3.31 | 0.581 | 0.235 | -0.475 | 0.476 | 0.791 | -0.197 | 7.6 | 1010 | 10 | -- | -- |
| 217.95 | 0.693 | 3.31 | 0.528 | 0.238 | -0.499 | 0.378 | -- | -0.206 | 7.43 | 1010 | 10 | -- | -- |
| 216.58 | 0.691 | 3.31 | 0.588 | 0.237 | -0.472 | -- | 0.737 | -0.198 | 7.53 | 1010 | 10 | -- | -- |
| 218.34 | 0.693 | 3.31 | 0.536 | 0.239 | -0.495 | -- | -- | -0.206 | 7.46 | 1010 | 10 | -- | -- |
| 219.52 | 0.694 | 3.31 | 0.594 | -- | -0.515 | -- | 0.749 | -0.195 | -- | 1010 | 10 | -- | -- |
| 225.38 | 0.704 | 3.31 | 0.507 | 0.29 | -- | 0.315 | -- | -0.205 | 7.72 | -- | -- | -- | -- |
| 225.66 | 0.703 | 3.31 | 0.515 | 0.29 | -- | -- | -- | -0.205 | 7.72 | -- | -- | -- | -- |
| 221.34 | 0.696 | 3.32 | 0.542 | -- | -0.538 | -- | -- | -0.202 | -- | 1010 | 9 | -- | -- |
| 225.91 | 0.704 | 3.33 | -- | 0.246 | -0.474 | -- | -- | -0.266 | 7.33 | 1010 | 9 | -- | -- |
| 230.2 | 0.709 | 3.32 | 0.519 | -- | -- | -- | -- | -0.201 | -- | -- | -- | -- | -- |
| 232.65 | 0.714 | 3.32 | -- | 0.294 | -- | -- | -- | -0.263 | 7.59 | -- | -- | -- | -- |
| 229.07 | 0.707 | 3.33 | -- | -- | -0.518 | -- | -- | -0.262 | -- | 1010 | 9 | -- | -- |
| 237.3 | 0.719 | 3.33 | -- | -- | -- | -- | -- | -0.259 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 204.02 | wind | 7 | 30 | > th | dose dependent | rect. | 1 | 9 |
| 6 | 204.11 | wind | 7 | 30 | > th | dose dependent | rect. | 4 | 6 |
| 11 | 204.14 | wind | 7 | 30 | > th | dose dependent | rect. | 7 | 3 |
| 25 | 204.27 | wind | 7 | 30 | > th | dose dependent | rect. | 9 | 1 |
| 40 | 204.80 | wind | 7 | 30 | > th | dose independent | rect. | 1 | 10 |
| 52 | 204.96 | wind | 7 | 30 | > th | dose independent | rect. | 7 | 4 |
| 56 | 204.99 | wind | 5 | 10 | > th | dose independent | rect. | 10 | 1 |
| 78 | 205.07 | wind | 9 | 10 | > th | dose independent | rect. | 19 | 16 |
| 81 | 205.10 | humidity | 60 | 10 | < th | dose independent | rect. | 10 | 1 |
| 85 | 205.10 | humidity | 60 | 10 | < th | dose dependent | rect. | 10 | 1 |
| 346 | 209.81 | atmosphere | 1010 | 10 | < th | dose independent | rect. | 4 | 23 |
| 371 | 211.27 | atmosphere | 1010 | 10 | < th | dose independent | rect. | 4 | 15 |
| 390 | 211.77 | atmosphere | 1010 | 10 | < th | dose independent | rect. | 4 | 12 |
| 393 | 212.19 | temperature | 25 | 30 | > th | dose dependent | rect. | 5 | 1 |
| 396 | 212.37 | atmosphere | 1005 | 10 | > th | dose dependent | rect. | 3 | 1 |
| 405 | 212.67 | atmosphere | 1005 | 10 | > th | dose independent | rect. | 3 | 1 |
| 406 | 212.68 | wind | 9 | 10 | > th | dose independent | sin | 7 | NA |
| 408 | 212.89 | atmosphere | 1010 | 10 | < th | dose independent | sin | 1 | NA |
| 414 | 213.01 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 5 | 1 |
| 421 | 213.01 | atmosphere | 1000 | 10 | < th | dose dependent | rect. | 5 | 1 |
| 429 | 213.03 | atmosphere | 1010 | 270 | > th | dose dependent | rect. | 10 | 1 |
| 433 | 213.09 | atmosphere | 1010 | 10 | > th | dose independent | rect. | 11 | 1 |
| 504 | 213.17 | wind | 9 | 10 | < th | dose independent | sin | 7 | NA |
| 558 | 213.61 | atmosphere | 1010 | 10 | > th | dose dependent | rect. | 11 | 1 |
| 730 | 214.03 | temperature | 10 | 10 | > th | dose dependent | rect. | 4 | 23 |
| 732 | 214.04 | atmosphere | 1010 | 270 | > th | dose dependent | sin | 1 | NA |
| 873 | 214.21 | atmosphere | 1010 | 10 | > th | dose dependent | sin | 23 | NA |
| 883 | 214.21 | wind | 9 | 43200 | > th | dose dependent | rect. | 18 | 14 |