Os10g0442700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Protein serine/threonine kinase (Protein kinase).

FiT-DB / Search/ Help/ Sample detail
|
Os10g0442700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Protein serine/threonine kinase (Protein kinase).
|
|
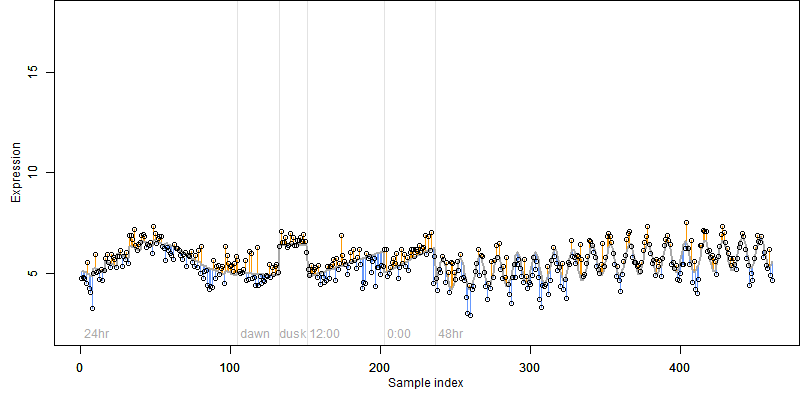
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

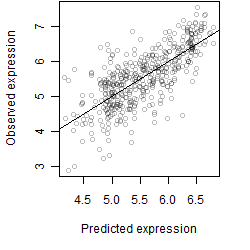 __
__

Dependence on each variable
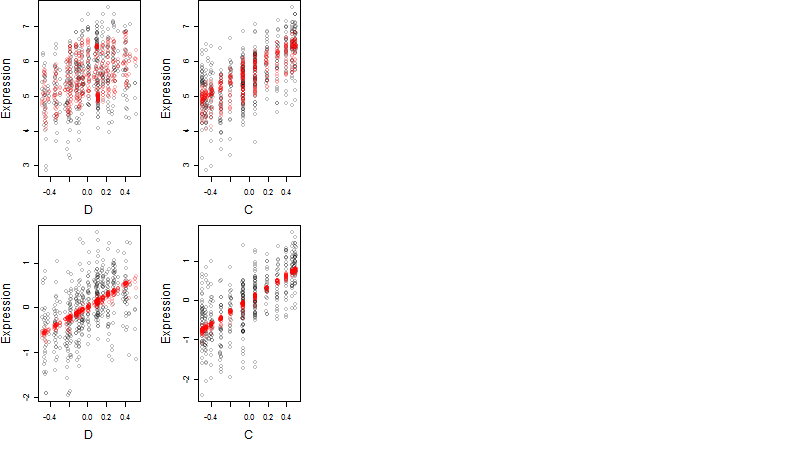
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 119.8 | 0.514 | 5.43 | 2.05 | 1.55 | -0.315 | 0.496 | -3.19 | 0.481 | 18.3 | 24.7 | 2328 | -- | -- |
| 129.13 | 0.533 | 5.52 | 1.17 | 1.44 | -0.394 | 0.676 | -- | 0.311 | 19 | 28.8 | 6186 | -- | -- |
| 120.39 | 0.511 | 5.43 | 2.06 | 1.55 | -0.311 | -- | -3.24 | 0.486 | 18.3 | 24.63 | 2296 | -- | -- |
| 130.3 | 0.535 | 5.52 | 1.16 | 1.45 | -0.393 | -- | -- | 0.313 | 19 | 28.8 | 6186 | -- | -- |
| 238.24 | 0.719 | 5.41 | 2.03 | -- | -0.593 | -- | -3.05 | 0.364 | -- | 25.3 | 2032 | -- | -- |
| 137.22 | 0.549 | 5.51 | 1.26 | 1.56 | -- | 0.705 | -- | 0.341 | 18.5 | -- | -- | -- | -- |
| 138.44 | 0.551 | 5.51 | 1.25 | 1.56 | -- | -- | -- | 0.343 | 18.5 | -- | -- | -- | -- |
| 235.98 | 0.719 | 5.49 | 1.09 | -- | -0.634 | -- | -- | 0.271 | -- | 28.8 | 6186 | -- | -- |
| 132.39 | 0.536 | 5.62 | -- | 1.56 | -1.01 | -- | -- | -0.147 | 18.4 | 22.09 | 47520 | -- | -- |
| 264.98 | 0.761 | 5.48 | 1.21 | -- | -- | -- | -- | 0.311 | -- | -- | -- | -- | -- |
| 179.72 | 0.627 | 5.54 | -- | 1.54 | -- | -- | -- | 0.204 | 18.5 | -- | -- | -- | -- |
| 259.53 | 0.75 | 5.59 | -- | -- | -0.974 | -- | -- | -0.164 | -- | 22.01 | 47520 | -- | -- |
| 303.74 | 0.813 | 5.51 | -- | -- | -- | -- | -- | 0.176 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance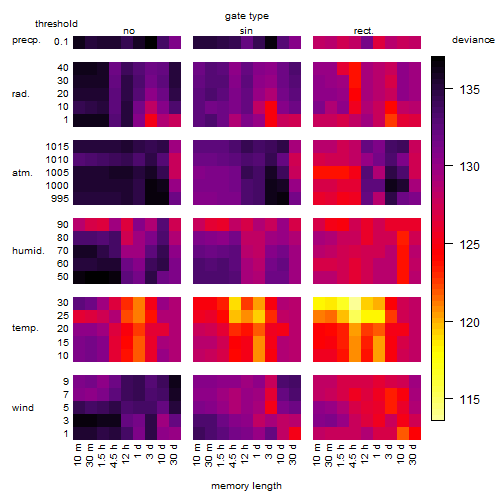
|
Histogram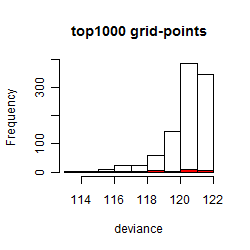 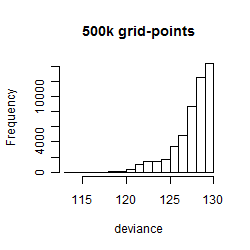
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 113.59 | temperature | 30 | 270 | < th | dose dependent | rect. | 6 | 9 |
| 6 | 115.12 | temperature | 25 | 270 | < th | dose independent | rect. | 6 | 7 |
| 25 | 116.35 | temperature | 30 | 270 | < th | dose dependent | rect. | 7 | 3 |
| 52 | 117.56 | temperature | 30 | 270 | < th | dose dependent | rect. | 9 | 1 |
| 73 | 118.18 | temperature | 25 | 1440 | < th | dose dependent | rect. | 5 | 6 |
| 76 | 118.24 | temperature | 30 | 10 | < th | dose dependent | rect. | 8 | 5 |
| 100 | 118.69 | temperature | 30 | 270 | < th | dose dependent | rect. | 6 | 15 |
| 111 | 118.89 | temperature | 30 | 270 | < th | dose dependent | sin | 1 | NA |
| 117 | 118.95 | temperature | 30 | 10 | < th | dose dependent | rect. | 8 | 8 |
| 136 | 119.09 | temperature | 25 | 1440 | < th | dose dependent | sin | 3 | NA |
| 217 | 119.76 | temperature | 25 | 270 | < th | dose independent | sin | 1 | NA |
| 235 | 119.86 | temperature | 25 | 270 | < th | dose independent | rect. | 6 | 15 |
| 314 | 120.23 | temperature | 10 | 1440 | > th | dose dependent | rect. | 4 | 6 |
| 331 | 120.28 | temperature | 10 | 1440 | > th | dose dependent | rect. | 2 | 10 |
| 370 | 120.40 | temperature | 25 | 720 | < th | dose independent | rect. | 23 | 13 |
| 392 | 120.45 | temperature | 20 | 270 | > th | dose dependent | rect. | 2 | 14 |
| 418 | 120.53 | temperature | 25 | 270 | > th | dose independent | rect. | 3 | 14 |
| 482 | 120.70 | temperature | 10 | 270 | > th | dose dependent | rect. | 2 | 14 |
| 537 | 120.83 | temperature | 10 | 1440 | > th | dose dependent | sin | 6 | NA |
| 598 | 120.91 | temperature | 25 | 1440 | < th | dose dependent | rect. | 14 | 23 |
| 602 | 120.93 | temperature | 25 | 1440 | < th | dose dependent | no | NA | NA |
| 730 | 121.10 | temperature | 25 | 10 | < th | dose independent | rect. | 8 | 8 |
| 756 | 121.13 | temperature | 10 | 270 | > th | dose dependent | rect. | 10 | 22 |
| 929 | 121.39 | temperature | 25 | 1440 | < th | dose independent | rect. | 18 | 23 |
| 939 | 121.40 | temperature | 25 | 1440 | < th | dose independent | rect. | 13 | 23 |
| 967 | 121.44 | temperature | 25 | 1440 | < th | dose independent | rect. | 23 | 22 |
| 981 | 121.46 | temperature | 25 | 1440 | < th | dose independent | rect. | 8 | 23 |