Os10g0435400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : TRAF-like domain containing protein.

FiT-DB / Search/ Help/ Sample detail
|
Os10g0435400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : TRAF-like domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
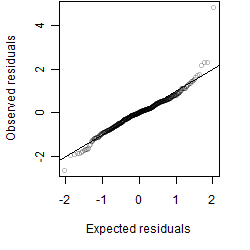
Dependence on each variable

Residual plot

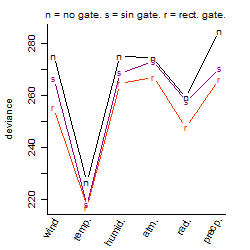
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 225.01 | 0.705 | 4.43 | -1.48 | 0.923 | -1.88 | -1.59 | -0.933 | -0.0506 | 21.1 | 23.4 | 2623 | -- | -- |
| 225.53 | 0.699 | 4.46 | -1.84 | 0.988 | -1.92 | -1.38 | -- | -0.115 | 21.2 | 23.5 | 2829 | -- | -- |
| 231.08 | 0.708 | 4.45 | -1.62 | 1 | -1.9 | -- | -0.582 | -0.1 | 21.5 | 23.4 | 2995 | -- | -- |
| 232.18 | 0.71 | 4.47 | -1.81 | 1.01 | -1.9 | -- | -- | -0.114 | 21.5 | 23.52 | 2938 | -- | -- |
| 274.95 | 0.772 | 4.41 | -1.78 | -- | -1.96 | -- | 0.124 | 0.00886 | -- | 24.2 | 2215 | -- | -- |
| 365.84 | 0.897 | 4.28 | -0.998 | 1.04 | -- | -1.22 | -- | 0.694 | 21.8 | -- | -- | -- | -- |
| 371.35 | 0.902 | 4.28 | -0.954 | 1.04 | -- | -- | -- | 0.695 | 22 | -- | -- | -- | -- |
| 274.33 | 0.771 | 4.41 | -1.78 | -- | -2.07 | -- | -- | -0.0229 | -- | 24.2 | 2215 | -- | -- |
| 307 | 0.816 | 4.37 | -- | 0.908 | -1.42 | -- | -- | 0.298 | 21.6 | 22.82 | 2214 | -- | -- |
| 436.82 | 0.977 | 4.25 | -0.941 | -- | -- | -- | -- | 0.696 | -- | -- | -- | -- | -- |
| 395.33 | 0.93 | 4.26 | -- | 1.04 | -- | -- | -- | 0.801 | 21.9 | -- | -- | -- | -- |
| 341.99 | 0.861 | 4.35 | -- | -- | -1.61 | -- | -- | 0.261 | -- | 24.84 | 841 | -- | -- |
| 460.19 | 1 | 4.23 | -- | -- | -- | -- | -- | 0.8 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 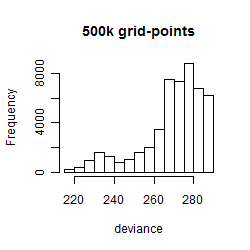
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 217.02 | temperature | 10 | 1440 | > th | dose dependent | rect. | 21 | 14 |
| 11 | 217.35 | temperature | 10 | 1440 | > th | dose dependent | rect. | 1 | 9 |
| 16 | 217.38 | temperature | 30 | 1440 | < th | dose dependent | rect. | 3 | 6 |
| 18 | 217.41 | temperature | 30 | 1440 | < th | dose dependent | rect. | 21 | 14 |
| 24 | 217.48 | temperature | 10 | 1440 | > th | dose dependent | rect. | 3 | 6 |
| 29 | 217.50 | temperature | 30 | 1440 | < th | dose dependent | rect. | 1 | 9 |
| 63 | 218.04 | temperature | 10 | 1440 | > th | dose dependent | sin | 8 | NA |
| 109 | 218.75 | temperature | 30 | 1440 | < th | dose dependent | sin | 8 | NA |
| 163 | 219.63 | temperature | 10 | 1440 | > th | dose dependent | rect. | 6 | 1 |
| 174 | 219.90 | temperature | 30 | 1440 | < th | dose dependent | rect. | 6 | 1 |
| 699 | 225.78 | temperature | 10 | 4320 | > th | dose dependent | rect. | 21 | 2 |
| 700 | 225.78 | temperature | 30 | 4320 | < th | dose dependent | rect. | 21 | 2 |
| 778 | 226.54 | temperature | 10 | 1440 | > th | dose dependent | no | NA | NA |
| 860 | 227.04 | temperature | 10 | 1440 | > th | dose dependent | rect. | 23 | 23 |
| 864 | 227.07 | temperature | 30 | 1440 | < th | dose dependent | no | NA | NA |
| 917 | 227.35 | temperature | 10 | 1440 | > th | dose dependent | rect. | 3 | 23 |
| 966 | 227.58 | temperature | 30 | 1440 | < th | dose dependent | rect. | 24 | 23 |
| 995 | 227.69 | temperature | 30 | 1440 | < th | dose dependent | rect. | 8 | 23 |
| 1000 | 227.72 | temperature | 30 | 1440 | < th | dose dependent | rect. | 3 | 23 |