Os10g0416200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Beta-ketoacyl-CoA synthase.


FiT-DB / Search/ Help/ Sample detail
|
Os10g0416200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Beta-ketoacyl-CoA synthase.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

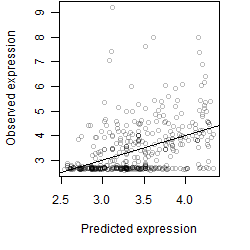
Dependence on each variable

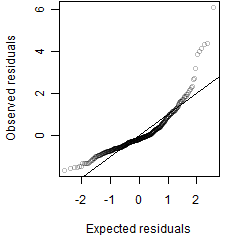
Residual plot
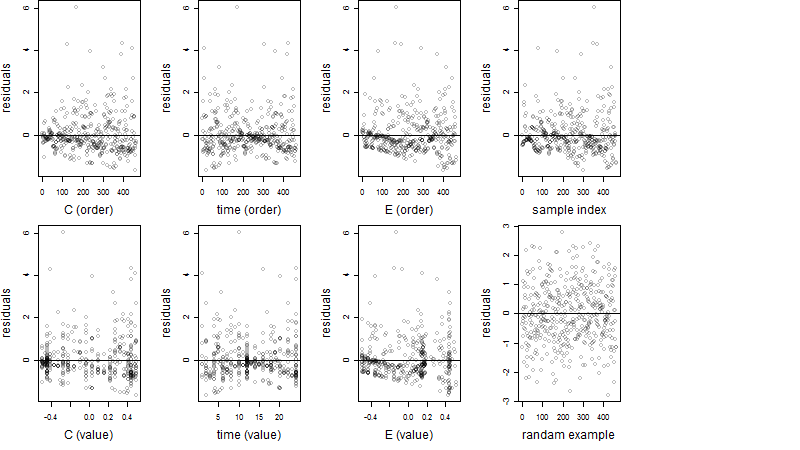
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 376.52 | 0.912 | 3.44 | 0.0125 | 0.699 | 1.78 | -0.464 | 1.86 | -0.445 | 1.61 | 8.989 | 1533 | -- | -- |
| 379.55 | 0.907 | 3.5 | -0.364 | 0.741 | 1.76 | -0.44 | -- | -0.517 | 1.68 | 15.68 | 1562 | -- | -- |
| 377.35 | 0.905 | 3.44 | 0.0238 | 0.708 | 1.78 | -- | 1.85 | -0.442 | 1.69 | 9.804 | 1553 | -- | -- |
| 379.11 | 0.907 | 3.5 | -0.298 | 0.737 | 1.34 | -- | -- | -0.523 | 1.9 | 20.21 | 1585 | -- | -- |
| 398.32 | 0.93 | 3.43 | -0.0647 | -- | 1.69 | -- | 1.49 | -0.467 | -- | 16.7 | 2486 | -- | -- |
| 434.11 | 0.977 | 3.42 | 0.177 | 0.748 | -- | -0.362 | -- | -0.222 | 1.66 | -- | -- | -- | -- |
| 434.63 | 0.976 | 3.42 | 0.188 | 0.749 | -- | -- | -- | -0.22 | 1.7 | -- | -- | -- | -- |
| 397.86 | 0.929 | 3.46 | -0.279 | -- | 1.41 | -- | -- | -0.444 | -- | 23.4 | 2541 | -- | -- |
| 380.63 | 0.909 | 3.47 | -- | 0.703 | 1.18 | -- | -- | -0.469 | 1.76 | 21.91 | 1531 | -- | -- |
| 468.77 | 1.01 | 3.41 | 0.219 | -- | -- | -- | -- | -0.203 | -- | -- | -- | -- | -- |
| 435.56 | 0.976 | 3.42 | -- | 0.752 | -- | -- | -- | -0.241 | 1.7 | -- | -- | -- | -- |
| 399.76 | 0.931 | 3.45 | -- | -- | 1.34 | -- | -- | -0.393 | -- | 23.56 | 2526 | -- | -- |
| 470.03 | 1.01 | 3.42 | -- | -- | -- | -- | -- | -0.228 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 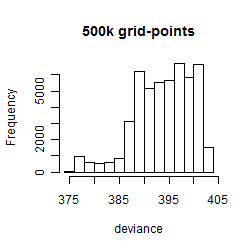
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 374.48 | temperature | 10 | 720 | > th | dose dependent | rect. | 12 | 13 |
| 2 | 374.85 | temperature | 10 | 720 | > th | dose dependent | rect. | 13 | 11 |
| 3 | 375.61 | temperature | 10 | 1440 | > th | dose dependent | rect. | 2 | 17 |
| 32 | 375.90 | temperature | 10 | 1440 | > th | dose dependent | sin | 1 | NA |
| 46 | 375.97 | temperature | 10 | 1440 | > th | dose dependent | rect. | 10 | 9 |
| 47 | 375.97 | temperature | 10 | 1440 | > th | dose dependent | rect. | 5 | 14 |
| 55 | 376.00 | temperature | 30 | 1440 | < th | dose dependent | rect. | 6 | 13 |
| 78 | 376.07 | temperature | 10 | 720 | > th | dose dependent | rect. | 12 | 17 |
| 102 | 376.17 | temperature | 10 | 1440 | > th | dose dependent | rect. | 8 | 19 |
| 155 | 376.30 | temperature | 30 | 1440 | < th | dose dependent | sin | 22 | NA |
| 157 | 376.30 | temperature | 30 | 1440 | < th | dose dependent | rect. | 2 | 17 |
| 312 | 376.60 | temperature | 10 | 1440 | > th | dose dependent | rect. | 10 | 5 |
| 417 | 376.74 | temperature | 30 | 1440 | < th | dose dependent | rect. | 15 | 4 |
| 494 | 376.85 | temperature | 30 | 1440 | < th | dose dependent | rect. | 6 | 19 |
| 529 | 376.89 | temperature | 10 | 1440 | > th | dose dependent | no | NA | NA |
| 559 | 376.93 | temperature | 30 | 720 | < th | dose dependent | rect. | 15 | 6 |
| 658 | 377.09 | temperature | 10 | 1440 | > th | dose dependent | rect. | 12 | 23 |
| 710 | 377.17 | temperature | 30 | 1440 | < th | dose dependent | rect. | 13 | 7 |
| 981 | 377.86 | temperature | 30 | 1440 | < th | dose dependent | no | NA | NA |