Os10g0416000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Non-protein coding transcript, unclassifiable transcript.

FiT-DB / Search/ Help/ Sample detail
|
Os10g0416000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Non-protein coding transcript, unclassifiable transcript.
|
|
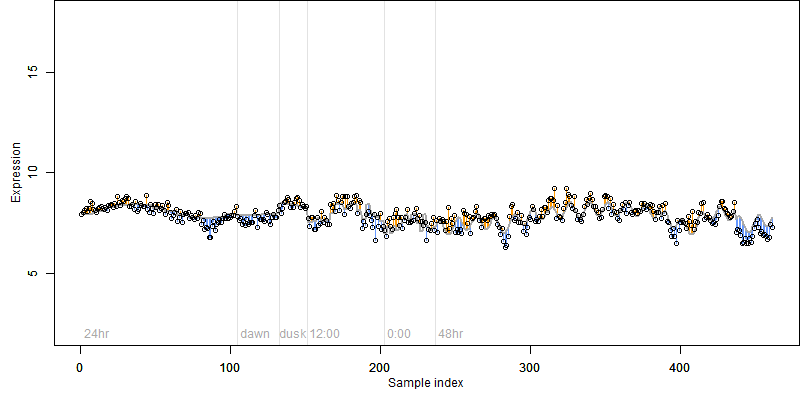
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
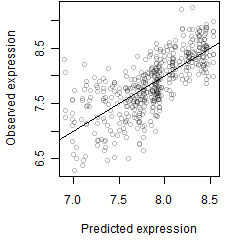
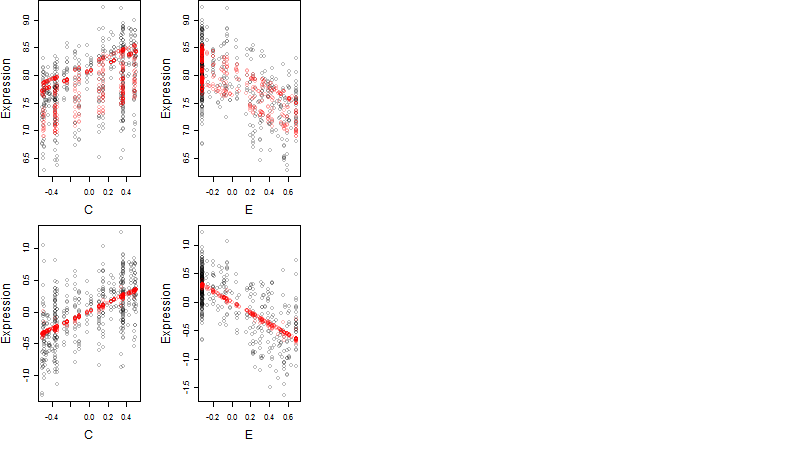
Dependence on each variable
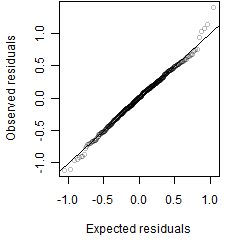
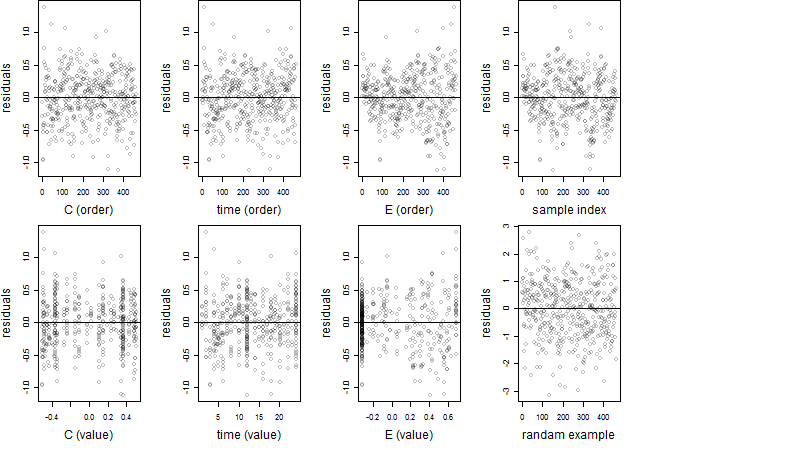
Residual plot
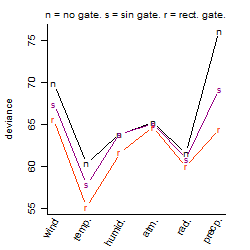
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 56.94 | 0.355 | 7.95 | -0.789 | 0.647 | -1.19 | 0.329 | 0.853 | -0.284 | 14.9 | 24.3 | 1582 | -- | -- |
| 56.63 | 0.35 | 7.93 | -0.497 | 0.717 | -1.08 | 0.26 | -- | -0.247 | 14.8 | 23.7 | 1461 | -- | -- |
| 56.6 | 0.35 | 7.95 | -0.797 | 0.724 | -1.11 | -- | 0.835 | -0.309 | 14.8 | 23.89 | 1470 | -- | -- |
| 56.99 | 0.352 | 7.92 | -0.469 | 0.687 | -1.06 | -- | -- | -0.221 | 14.9 | 23.57 | 1474 | -- | -- |
| 80.38 | 0.418 | 7.95 | -0.798 | -- | -1.22 | -- | 0.896 | -0.33 | -- | 24.3 | 1654 | -- | -- |
| 105.63 | 0.482 | 7.81 | -0.0506 | 0.634 | -- | 0.334 | -- | 0.255 | 14.9 | -- | -- | -- | -- |
| 106 | 0.482 | 7.81 | -0.0431 | 0.635 | -- | -- | -- | 0.257 | 14.9 | -- | -- | -- | -- |
| 81.83 | 0.421 | 7.94 | -0.517 | -- | -1.23 | -- | -- | -0.252 | -- | 24.3 | 1658 | -- | -- |
| 62.24 | 0.367 | 7.9 | -- | 0.695 | -0.945 | -- | -- | -0.125 | 14.9 | 23.59 | 1445 | -- | -- |
| 129.25 | 0.531 | 7.81 | -0.0702 | -- | -- | -- | -- | 0.24 | -- | -- | -- | -- | -- |
| 106.05 | 0.482 | 7.81 | -- | 0.636 | -- | -- | -- | 0.262 | 14.9 | -- | -- | -- | -- |
| 87.53 | 0.437 | 7.89 | -- | -- | -0.689 | -- | -- | -0.0996 | -- | 24.3 | 1658 | -- | -- |
| 129.38 | 0.531 | 7.81 | -- | -- | -- | -- | -- | 0.248 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance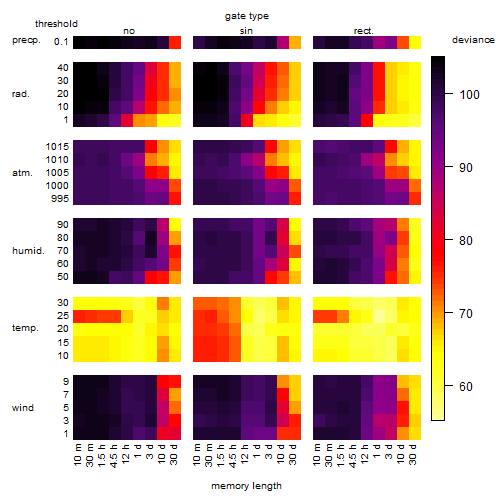
|
Histogram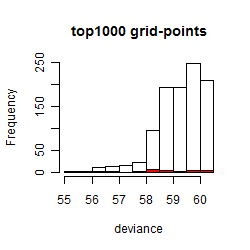 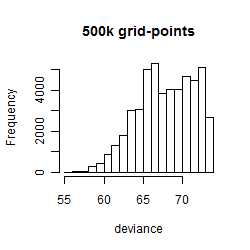
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 55.20 | temperature | 25 | 1440 | < th | dose independent | rect. | 15 | 7 |
| 3 | 55.89 | temperature | 25 | 1440 | > th | dose independent | rect. | 14 | 8 |
| 13 | 56.48 | temperature | 25 | 1440 | > th | dose independent | rect. | 11 | 11 |
| 14 | 56.68 | temperature | 25 | 1440 | < th | dose independent | rect. | 11 | 12 |
| 55 | 57.88 | temperature | 25 | 1440 | < th | dose independent | sin | 19 | NA |
| 61 | 57.99 | temperature | 25 | 4320 | < th | dose dependent | rect. | 23 | 4 |
| 67 | 58.04 | temperature | 25 | 1440 | > th | dose independent | sin | 20 | NA |
| 73 | 58.07 | temperature | 25 | 4320 | < th | dose dependent | rect. | 1 | 2 |
| 78 | 58.08 | temperature | 25 | 4320 | < th | dose dependent | rect. | 21 | 9 |
| 114 | 58.31 | temperature | 30 | 1440 | < th | dose dependent | rect. | 14 | 16 |
| 127 | 58.40 | temperature | 10 | 4320 | > th | dose dependent | rect. | 1 | 2 |
| 146 | 58.48 | temperature | 15 | 4320 | > th | dose dependent | rect. | 23 | 7 |
| 190 | 58.59 | temperature | 20 | 1440 | > th | dose dependent | rect. | 15 | 15 |
| 226 | 58.65 | temperature | 30 | 1440 | < th | dose dependent | sin | 15 | NA |
| 283 | 58.79 | temperature | 20 | 720 | > th | dose dependent | rect. | 21 | 18 |
| 336 | 58.96 | temperature | 20 | 1440 | > th | dose dependent | sin | 14 | NA |
| 390 | 59.14 | temperature | 25 | 4320 | < th | dose dependent | sin | 11 | NA |
| 481 | 59.32 | temperature | 30 | 720 | < th | dose dependent | rect. | 1 | 18 |
| 632 | 59.70 | temperature | 30 | 1440 | < th | dose dependent | rect. | 16 | 3 |
| 655 | 59.74 | temperature | 20 | 720 | > th | dose dependent | sin | 5 | NA |
| 768 | 59.97 | radiation | 1 | 43200 | < th | dose dependent | rect. | 17 | 2 |
| 810 | 60.03 | radiation | 1 | 43200 | < th | dose dependent | rect. | 17 | 11 |
| 847 | 60.10 | radiation | 1 | 43200 | < th | dose independent | rect. | 17 | 2 |
| 848 | 60.10 | radiation | 1 | 43200 | < th | dose independent | rect. | 17 | 4 |
| 860 | 60.11 | radiation | 1 | 43200 | < th | dose independent | rect. | 17 | 11 |
| 980 | 60.30 | temperature | 20 | 1440 | > th | dose dependent | rect. | 20 | 1 |