Os10g0410900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Hypothetical protein.


FiT-DB / Search/ Help/ Sample detail
|
Os10g0410900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
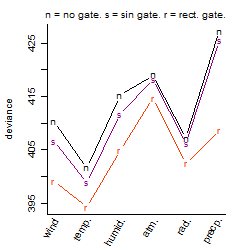
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 400.62 | 0.94 | 4.12 | -1.25 | 0.0366 | 1.26 | -1.01 | -1.53 | -0.37 | 15.2 | 20.71 | 1306 | -- | -- |
| 402.59 | 0.934 | 4.07 | -0.814 | 0.0523 | 1.2 | -0.913 | -- | -0.248 | 15.1 | 20.3 | 1269 | -- | -- |
| 403.74 | 0.936 | 4.12 | -1.28 | 0.0306 | 1.26 | -- | -1.55 | -0.379 | 16.2 | 21.21 | 1335 | -- | -- |
| 405.16 | 0.937 | 4.07 | -0.83 | 0.0486 | 1.19 | -- | -- | -0.253 | 15.3 | 20.54 | 1263 | -- | -- |
| 403.75 | 0.936 | 4.12 | -1.26 | -- | 1.22 | -- | -1.47 | -0.371 | -- | 21.2 | 1360 | -- | -- |
| 444.3 | 0.988 | 4.01 | -0.391 | 0.12 | -- | 0.967 | -- | 0.0205 | 2.54 | -- | -- | -- | -- |
| 447.41 | 0.991 | 4.01 | -0.416 | 0.133 | -- | -- | -- | 0.0155 | 0.614 | -- | -- | -- | -- |
| 405.25 | 0.938 | 4.07 | -0.827 | -- | 1.17 | -- | -- | -0.254 | -- | 20.57 | 1299 | -- | -- |
| 420.96 | 0.956 | 4.04 | -- | 0.05 | 1.03 | -- | -- | -0.114 | 15 | 21.36 | 1078 | -- | -- |
| 448.51 | 0.99 | 4.01 | -0.411 | -- | -- | -- | -- | 0.0178 | -- | -- | -- | -- | -- |
| 451.96 | 0.994 | 4 | -- | 0.126 | -- | -- | -- | 0.0618 | 0.455 | -- | -- | -- | -- |
| 421.03 | 0.956 | 4.04 | -- | -- | 0.984 | -- | -- | -0.109 | -- | 21.1 | 1071 | -- | -- |
| 452.96 | 0.993 | 4 | -- | -- | -- | -- | -- | 0.0634 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 394.24 | temperature | 20 | 1440 | > th | dose dependent | rect. | 17 | 2 |
| 6 | 395.15 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 1 |
| 19 | 396.09 | temperature | 30 | 1440 | < th | dose dependent | rect. | 17 | 2 |
| 35 | 396.77 | temperature | 30 | 720 | < th | dose dependent | rect. | 4 | 17 |
| 53 | 397.47 | temperature | 25 | 1440 | < th | dose independent | rect. | 14 | 8 |
| 70 | 397.63 | temperature | 25 | 1440 | < th | dose independent | rect. | 11 | 11 |
| 72 | 397.68 | temperature | 25 | 1440 | > th | dose independent | rect. | 11 | 11 |
| 77 | 397.70 | temperature | 30 | 720 | < th | dose dependent | rect. | 4 | 21 |
| 128 | 398.46 | temperature | 10 | 720 | > th | dose dependent | rect. | 16 | 23 |
| 158 | 398.85 | temperature | 20 | 1440 | > th | dose dependent | sin | 17 | NA |
| 202 | 399.16 | wind | 3 | 43200 | > th | dose dependent | rect. | 20 | 1 |
| 243 | 399.49 | temperature | 10 | 1440 | > th | dose dependent | sin | 18 | NA |
| 273 | 399.66 | temperature | 25 | 1440 | > th | dose independent | rect. | 15 | 5 |
| 309 | 399.86 | temperature | 10 | 720 | > th | dose dependent | rect. | 1 | 19 |
| 416 | 400.30 | temperature | 10 | 720 | > th | dose dependent | rect. | 1 | 21 |
| 443 | 400.39 | temperature | 30 | 720 | < th | dose dependent | sin | 0 | NA |
| 569 | 400.74 | temperature | 30 | 1440 | < th | dose dependent | sin | 18 | NA |
| 577 | 400.76 | wind | 9 | 43200 | < th | dose dependent | rect. | 20 | 1 |
| 621 | 400.88 | temperature | 25 | 1440 | < th | dose independent | sin | 20 | NA |
| 732 | 401.15 | temperature | 10 | 720 | > th | dose dependent | rect. | 23 | 21 |
| 733 | 401.16 | temperature | 25 | 1440 | > th | dose independent | sin | 21 | NA |
| 854 | 401.43 | temperature | 10 | 720 | > th | dose dependent | rect. | 21 | 22 |
| 921 | 401.59 | temperature | 10 | 720 | > th | dose dependent | rect. | 23 | 23 |
| 944 | 401.64 | temperature | 20 | 1440 | > th | dose dependent | no | NA | NA |
| 960 | 401.67 | wind | 5 | 43200 | < th | dose independent | rect. | 19 | 2 |