Os10g0375000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein kinase-like domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os10g0375000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein kinase-like domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

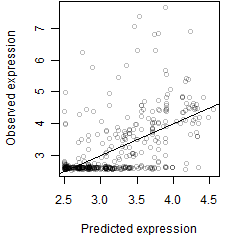 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 240.51 | 0.729 | 3.18 | -0.162 | 0.469 | 1.78 | -0.956 | -0.127 | -0.134 | 5.8 | 21.59 | 825 | 7.65 | -- |
| 240.52 | 0.722 | 3.18 | -0.137 | 0.472 | 1.77 | -0.959 | -- | -0.127 | 5.8 | 21.55 | 825 | 7.71 | -- |
| 242.62 | 0.725 | 3.18 | -0.212 | 0.48 | 1.78 | -- | -0.309 | -0.134 | 6.09 | 21.4 | 805 | 7.77 | -- |
| 242.72 | 0.726 | 3.18 | -0.141 | 0.486 | 1.76 | -- | -- | -0.116 | 6.17 | 21.4 | 819 | 7.91 | -- |
| 248.08 | 0.734 | 3.21 | -0.29 | -- | 2.1 | -- | -0.374 | -0.222 | -- | 19.99 | 673 | 6.37 | -- |
| 308.15 | 0.823 | 3.11 | 0.233 | 1.1 | -- | -0.449 | -- | 0.173 | 4.3 | -- | -- | -- | -- |
| 308.7 | 0.823 | 3.11 | 0.237 | 1.1 | -- | -- | -- | 0.176 | 4.27 | -- | -- | -- | -- |
| 248.11 | 0.734 | 3.21 | -0.178 | -- | 2.07 | -- | -- | -0.202 | -- | 20.06 | 675 | 6.33 | -- |
| 243.2 | 0.726 | 3.17 | -- | 0.496 | 1.72 | -- | -- | -0.0955 | 6.13 | 21.47 | 820 | 7.91 | -- |
| 374.05 | 0.904 | 3.11 | 0.281 | -- | -- | -- | -- | 0.205 | -- | -- | -- | -- | -- |
| 310.19 | 0.824 | 3.11 | -- | 1.11 | -- | -- | -- | 0.149 | 4.27 | -- | -- | -- | -- |
| 248.93 | 0.735 | 3.2 | -- | -- | 2.04 | -- | -- | -0.178 | -- | 20.09 | 667 | 6.28 | -- |
| 376.14 | 0.905 | 3.12 | -- | -- | -- | -- | -- | 0.174 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 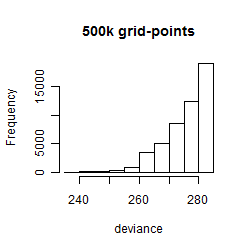
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 235.51 | temperature | 25 | 720 | > th | dose independent | rect. | 18 | 4 |
| 2 | 235.67 | temperature | 20 | 720 | > th | dose dependent | rect. | 18 | 9 |
| 11 | 237.80 | temperature | 10 | 720 | > th | dose dependent | rect. | 18 | 7 |
| 12 | 237.85 | temperature | 25 | 720 | < th | dose independent | rect. | 17 | 9 |
| 32 | 239.76 | temperature | 30 | 720 | < th | dose dependent | rect. | 17 | 9 |
| 36 | 240.09 | temperature | 10 | 720 | > th | dose dependent | rect. | 18 | 11 |
| 83 | 242.64 | temperature | 20 | 720 | > th | dose dependent | sin | 13 | NA |
| 104 | 243.67 | temperature | 20 | 270 | > th | dose dependent | rect. | 24 | 6 |
| 135 | 245.01 | temperature | 25 | 720 | < th | dose independent | sin | 15 | NA |
| 148 | 245.57 | temperature | 30 | 720 | < th | dose dependent | sin | 15 | NA |
| 175 | 246.66 | temperature | 25 | 720 | > th | dose independent | sin | 15 | NA |
| 211 | 247.85 | temperature | 10 | 720 | > th | dose dependent | sin | 14 | NA |
| 225 | 248.18 | temperature | 25 | 720 | < th | dose independent | rect. | 19 | 2 |
| 283 | 249.88 | temperature | 30 | 270 | < th | dose dependent | rect. | 19 | 15 |
| 296 | 250.22 | temperature | 20 | 10 | > th | dose dependent | rect. | 2 | 5 |
| 299 | 250.32 | temperature | 25 | 720 | < th | dose independent | rect. | 16 | 16 |
| 358 | 251.51 | temperature | 20 | 270 | > th | dose independent | rect. | 2 | 3 |
| 458 | 253.08 | temperature | 20 | 270 | > th | dose independent | rect. | 24 | 6 |
| 467 | 253.20 | temperature | 10 | 720 | > th | dose dependent | rect. | 13 | 16 |
| 479 | 253.34 | temperature | 10 | 720 | > th | dose dependent | rect. | 11 | 18 |
| 503 | 253.78 | temperature | 25 | 720 | > th | dose independent | rect. | 11 | 15 |
| 533 | 254.20 | temperature | 20 | 10 | > th | dose dependent | rect. | 24 | 7 |
| 535 | 254.22 | temperature | 20 | 30 | > th | dose independent | rect. | 2 | 5 |
| 657 | 255.68 | temperature | 20 | 10 | > th | dose dependent | rect. | 21 | 10 |
| 699 | 256.20 | temperature | 10 | 270 | > th | dose dependent | rect. | 9 | 22 |
| 860 | 257.83 | temperature | 20 | 10 | > th | dose independent | rect. | 4 | 3 |
| 1000 | 258.69 | temperature | 20 | 1440 | > th | dose dependent | sin | 9 | NA |