Os10g0372100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Hypothetical protein.

FiT-DB / Search/ Help/ Sample detail
|
Os10g0372100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Hypothetical protein.
|
|
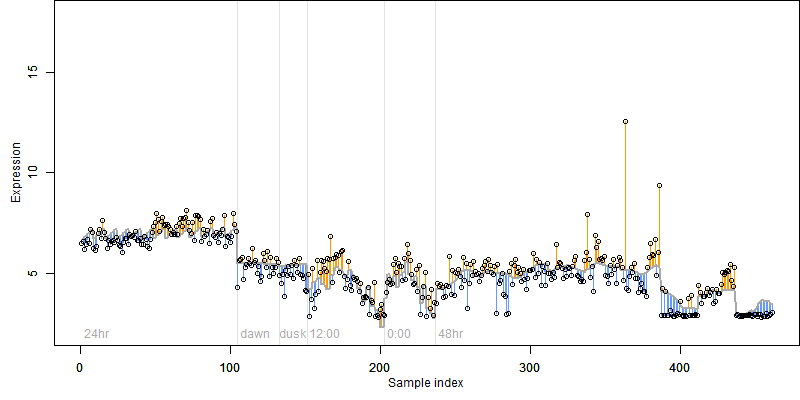
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
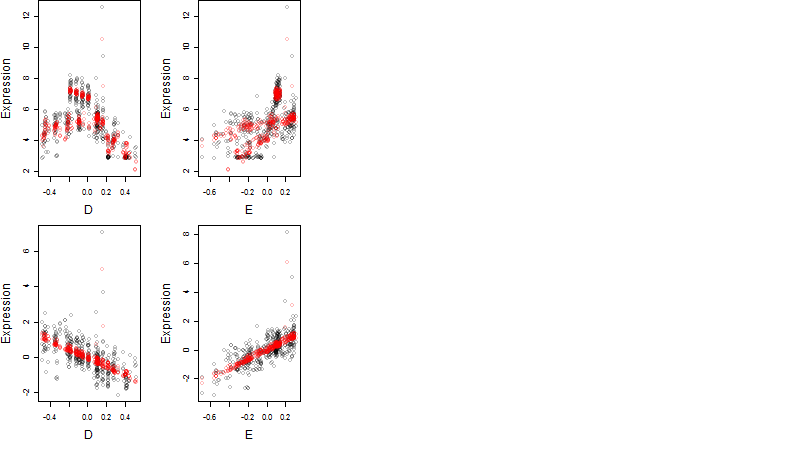
Dependence on each variable
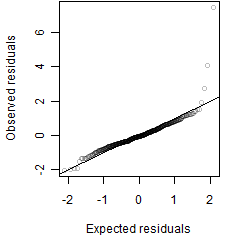
Residual plot
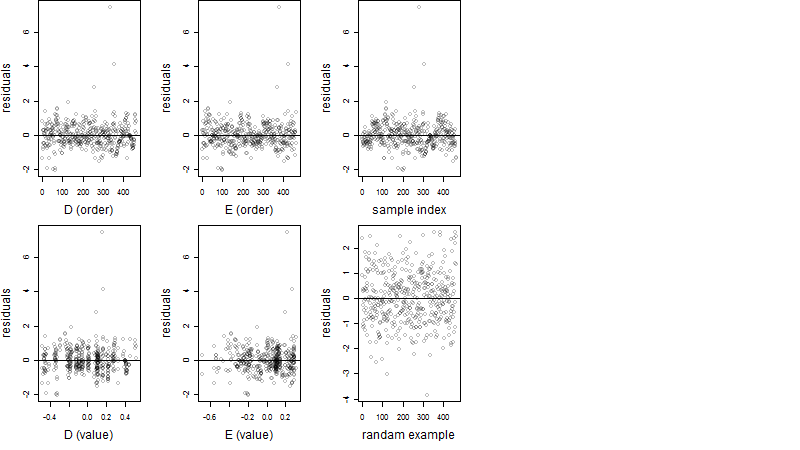
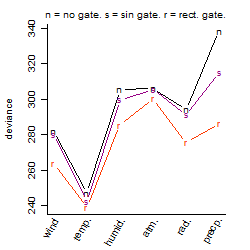
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 246.24 | 0.737 | 4.8 | -2.45 | 0.0403 | 3.2 | -0.733 | -0.167 | 1.6 | 18.2 | 6.064 | 1361 | -- | -- |
| 246.25 | 0.731 | 4.79 | -2.42 | 0.0426 | 3.2 | -0.728 | -- | 1.61 | 18.2 | 6.224 | 1352 | -- | -- |
| 246.63 | 0.731 | 4.8 | -2.42 | 0.162 | 3.15 | -- | -0.0691 | 1.61 | 23.7 | 4.75 | 1430 | -- | -- |
| 246.61 | 0.731 | 4.8 | -2.42 | 0.155 | 3.16 | -- | -- | 1.6 | 0.0328 | 2.68 | 1435 | -- | -- |
| 247.62 | 0.733 | 4.8 | -2.42 | -- | 3.19 | -- | -0.0675 | 1.6 | -- | 11.02 | 1355 | -- | -- |
| 416.95 | 0.957 | 4.66 | -1.4 | 0.3 | -- | 0.384 | -- | 2.19 | 1.88 | -- | -- | -- | -- |
| 417.46 | 0.957 | 4.67 | -1.41 | 0.3 | -- | -- | -- | 2.19 | 1.45 | -- | -- | -- | -- |
| 247.62 | 0.733 | 4.79 | -2.41 | -- | 3.19 | -- | -- | 1.61 | -- | 9.492 | 1355 | -- | -- |
| 368.89 | 0.895 | 4.7 | -- | 0.111 | 1.63 | -- | -- | 2.03 | 23.9 | 21.25 | 1380 | -- | -- |
| 422.98 | 0.961 | 4.66 | -1.4 | -- | -- | -- | -- | 2.19 | -- | -- | -- | -- | -- |
| 470.08 | 1.01 | 4.63 | -- | 0.275 | -- | -- | -- | 2.34 | 1.28 | -- | -- | -- | -- |
| 369.1 | 0.895 | 4.7 | -- | -- | 1.71 | -- | -- | 2.03 | -- | 21.2 | 1280 | -- | -- |
| 474.76 | 1.02 | 4.63 | -- | -- | -- | -- | -- | 2.35 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance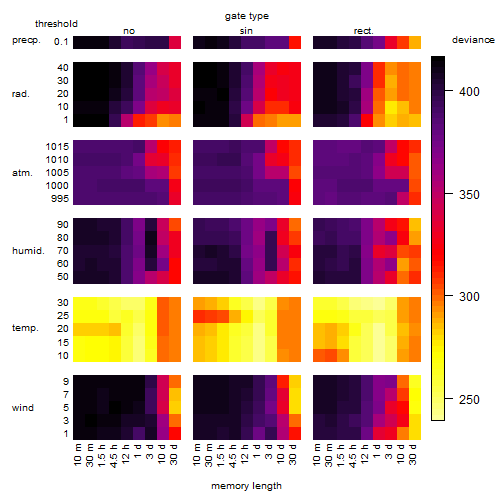
|
Histogram 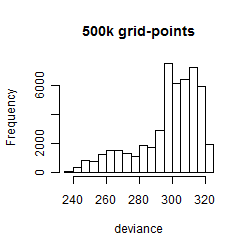
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 238.99 | temperature | 30 | 1440 | < th | dose dependent | rect. | 17 | 8 |
| 2 | 239.21 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 8 |
| 3 | 239.28 | temperature | 30 | 1440 | < th | dose dependent | rect. | 17 | 6 |
| 9 | 239.58 | temperature | 10 | 1440 | > th | dose dependent | rect. | 19 | 4 |
| 105 | 241.96 | temperature | 10 | 1440 | > th | dose dependent | sin | 15 | NA |
| 149 | 242.64 | temperature | 30 | 1440 | < th | dose dependent | sin | 15 | NA |
| 220 | 243.94 | temperature | 30 | 720 | < th | dose dependent | rect. | 10 | 21 |
| 238 | 244.16 | temperature | 10 | 1440 | > th | dose dependent | rect. | 11 | 17 |
| 393 | 245.57 | temperature | 10 | 1440 | > th | dose dependent | rect. | 16 | 20 |
| 407 | 245.67 | temperature | 30 | 1440 | < th | dose dependent | rect. | 10 | 17 |
| 421 | 245.81 | temperature | 30 | 1440 | < th | dose dependent | rect. | 16 | 20 |
| 602 | 246.66 | temperature | 30 | 720 | < th | dose dependent | rect. | 1 | 23 |
| 618 | 246.73 | temperature | 10 | 1440 | > th | dose dependent | rect. | 3 | 23 |
| 637 | 246.81 | temperature | 10 | 1440 | > th | dose dependent | no | NA | NA |
| 660 | 246.91 | temperature | 30 | 720 | < th | dose dependent | rect. | 5 | 22 |
| 716 | 247.15 | temperature | 30 | 720 | < th | dose dependent | rect. | 3 | 23 |
| 738 | 247.24 | temperature | 10 | 1440 | > th | dose dependent | rect. | 21 | 23 |
| 804 | 247.61 | temperature | 30 | 1440 | < th | dose dependent | no | NA | NA |
| 940 | 248.41 | temperature | 30 | 1440 | < th | dose dependent | rect. | 20 | 23 |