Os10g0349900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to NT16 polypeptide.

FiT-DB / Search/ Help/ Sample detail
|
Os10g0349900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to NT16 polypeptide.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
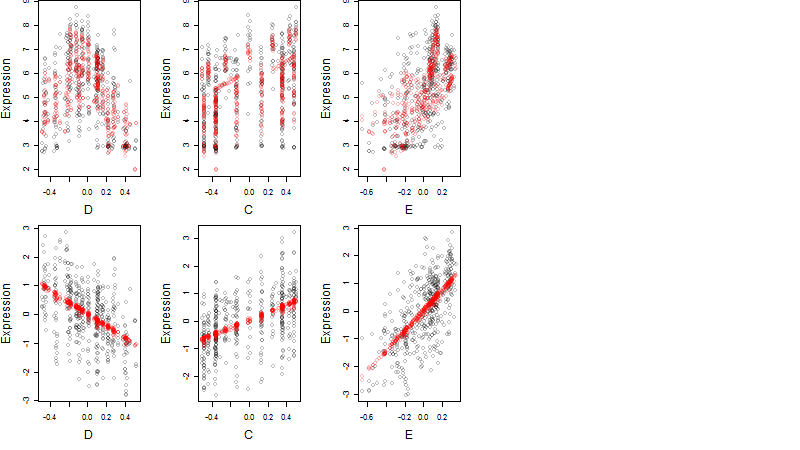
Dependence on each variable
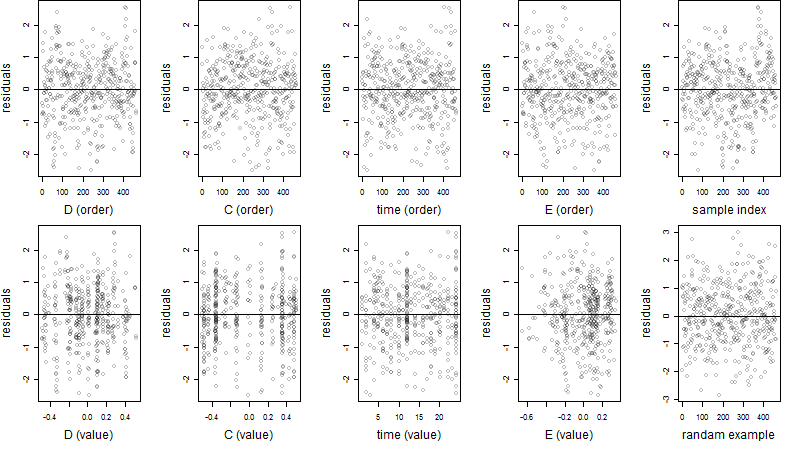
Residual plot
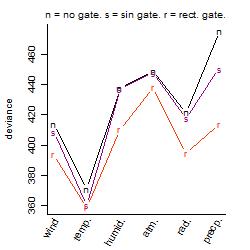
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 365.98 | 0.899 | 5.08 | -1.83 | 1.49 | 3.63 | -2.1 | 1.49 | 1.2 | 21 | 12.33 | 1223 | -- | -- |
| 366.83 | 0.892 | 5.11 | -2.15 | 1.22 | 3.8 | -2.09 | -- | 1.12 | 20.9 | 9.794 | 1052 | -- | -- |
| 380.95 | 0.909 | 5.09 | -1.88 | 1.47 | 3.64 | -- | 0.882 | 1.17 | 21 | 13.04 | 1196 | -- | -- |
| 381.43 | 0.91 | 5.11 | -2.07 | 1.39 | 3.69 | -- | -- | 1.12 | 21 | 8.415 | 1146 | -- | -- |
| 428.2 | 0.964 | 5.1 | -2.11 | -- | 4.72 | -- | 0.439 | 1.05 | -- | 14.69 | 820 | -- | -- |
| 584.66 | 1.13 | 4.98 | -1.04 | 1.71 | -- | -1.66 | -- | 1.76 | 21.8 | -- | -- | -- | -- |
| 595.11 | 1.14 | 4.98 | -0.979 | 1.71 | -- | -- | -- | 1.76 | 21.9 | -- | -- | -- | -- |
| 426.78 | 0.962 | 5.09 | -2.18 | -- | 4.39 | -- | -- | 1.03 | -- | 15.88 | 824 | -- | -- |
| 479.62 | 1.02 | 5.03 | -- | 1.37 | 2.85 | -- | -- | 1.48 | 21.1 | 11.35 | 1076 | -- | -- |
| 771.14 | 1.3 | 4.93 | -0.96 | -- | -- | -- | -- | 1.76 | -- | -- | -- | -- | -- |
| 620.38 | 1.17 | 4.95 | -- | 1.71 | -- | -- | -- | 1.87 | 21.8 | -- | -- | -- | -- |
| 525.2 | 1.07 | 5.01 | -- | -- | 3.05 | -- | -- | 1.4 | -- | 20.72 | 812 | -- | -- |
| 795.46 | 1.32 | 4.9 | -- | -- | -- | -- | -- | 1.87 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 359.11 | temperature | 10 | 1440 | > th | dose dependent | rect. | 15 | 9 |
| 2 | 359.15 | temperature | 10 | 1440 | > th | dose dependent | rect. | 14 | 12 |
| 9 | 359.35 | temperature | 20 | 1440 | > th | dose dependent | rect. | 13 | 14 |
| 17 | 359.50 | temperature | 20 | 1440 | > th | dose dependent | rect. | 13 | 16 |
| 52 | 360.42 | temperature | 10 | 1440 | > th | dose dependent | sin | 16 | NA |
| 160 | 362.85 | temperature | 30 | 1440 | < th | dose dependent | rect. | 15 | 8 |
| 263 | 365.09 | temperature | 30 | 1440 | < th | dose dependent | sin | 16 | NA |
| 555 | 370.11 | temperature | 30 | 720 | < th | dose dependent | rect. | 4 | 21 |
| 616 | 371.01 | temperature | 10 | 1440 | > th | dose dependent | no | NA | NA |
| 620 | 371.02 | temperature | 10 | 1440 | > th | dose dependent | rect. | 4 | 23 |
| 624 | 371.09 | temperature | 10 | 1440 | > th | dose dependent | rect. | 1 | 23 |
| 690 | 371.70 | temperature | 10 | 1440 | > th | dose dependent | rect. | 20 | 23 |