Os10g0345100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Multi antimicrobial extrusion protein MatE family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os10g0345100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Multi antimicrobial extrusion protein MatE family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

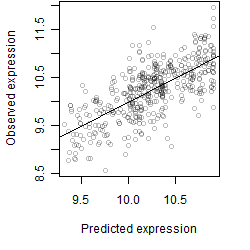 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 69.44 | 0.392 | 10.1 | 0.72 | 0.368 | -1.55 | 0.586 | 1.28 | 0.353 | 8.74 | 20.28 | 1261 | -- | -- |
| 70.26 | 0.39 | 10.2 | 0.494 | 0.342 | -1.58 | 0.615 | -- | 0.264 | 8.73 | 18.29 | 2569 | -- | -- |
| 70.52 | 0.391 | 10.1 | 0.714 | 0.367 | -1.55 | -- | 1.18 | 0.346 | 8.42 | 20.3 | 1251 | -- | -- |
| 70.85 | 0.392 | 10.2 | 0.545 | 0.345 | -1.71 | -- | -- | 0.287 | 8.6 | 16.53 | 2562 | -- | -- |
| 72.41 | 0.396 | 10.1 | 0.702 | -- | -1.72 | -- | 1.32 | 0.349 | -- | 20.51 | 958 | -- | -- |
| 134.37 | 0.543 | 10.2 | -0.164 | 0.466 | -- | 0.601 | -- | -0.0827 | 10.2 | -- | -- | -- | -- |
| 135.82 | 0.546 | 10.2 | -0.14 | 0.463 | -- | -- | -- | -0.0814 | 10.2 | -- | -- | -- | -- |
| 72.86 | 0.398 | 10.2 | 0.393 | -- | -1.74 | -- | -- | 0.258 | -- | 19.28 | 925 | -- | -- |
| 76.32 | 0.407 | 10.2 | -- | 0.292 | -1.33 | -- | -- | 0.157 | 8.27 | 20.84 | 2501 | -- | -- |
| 148.83 | 0.57 | 10.3 | -0.147 | -- | -- | -- | -- | -0.0823 | -- | -- | -- | -- | -- |
| 136.34 | 0.546 | 10.2 | -- | 0.464 | -- | -- | -- | -0.0658 | 10.2 | -- | -- | -- | -- |
| 76.17 | 0.406 | 10.2 | -- | -- | -1.58 | -- | -- | 0.188 | -- | 20.44 | 919 | -- | -- |
| 149.39 | 0.571 | 10.3 | -- | -- | -- | -- | -- | -0.066 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram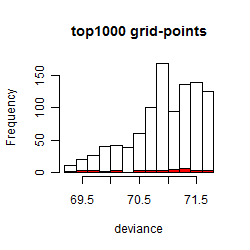 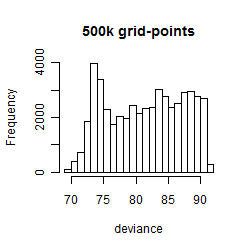
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 69.25 | temperature | 20 | 1440 | > th | dose dependent | rect. | 16 | 19 |
| 14 | 69.45 | temperature | 20 | 1440 | > th | dose dependent | sin | 7 | NA |
| 30 | 69.60 | temperature | 20 | 1440 | > th | dose dependent | rect. | 6 | 1 |
| 31 | 69.62 | temperature | 20 | 1440 | > th | dose dependent | rect. | 4 | 4 |
| 50 | 69.73 | temperature | 20 | 1440 | > th | dose dependent | rect. | 3 | 6 |
| 81 | 69.93 | temperature | 20 | 1440 | > th | dose dependent | no | NA | NA |
| 100 | 70.01 | temperature | 20 | 1440 | > th | dose dependent | rect. | 24 | 23 |
| 115 | 70.09 | temperature | 20 | 1440 | > th | dose dependent | rect. | 6 | 23 |
| 134 | 70.19 | temperature | 20 | 1440 | > th | dose dependent | rect. | 1 | 10 |
| 189 | 70.47 | temperature | 10 | 1440 | > th | dose dependent | rect. | 4 | 5 |
| 224 | 70.57 | temperature | 10 | 1440 | > th | dose dependent | rect. | 2 | 9 |
| 226 | 70.58 | temperature | 30 | 1440 | < th | dose dependent | rect. | 4 | 5 |
| 242 | 70.62 | temperature | 10 | 1440 | > th | dose dependent | rect. | 3 | 7 |
| 294 | 70.75 | temperature | 10 | 1440 | > th | dose dependent | rect. | 5 | 3 |
| 375 | 70.85 | temperature | 20 | 43200 | > th | dose independent | rect. | 19 | 2 |
| 380 | 70.86 | temperature | 30 | 1440 | < th | dose dependent | rect. | 5 | 3 |
| 433 | 70.91 | temperature | 10 | 1440 | > th | dose dependent | rect. | 6 | 1 |
| 506 | 71.01 | radiation | 1 | 14400 | > th | dose independent | rect. | 5 | 2 |
| 507 | 71.01 | radiation | 1 | 14400 | < th | dose independent | rect. | 5 | 2 |
| 546 | 71.10 | temperature | 30 | 1440 | < th | dose dependent | rect. | 6 | 1 |
| 596 | 71.20 | temperature | 30 | 4320 | < th | dose dependent | rect. | 21 | 12 |
| 602 | 71.21 | temperature | 10 | 4320 | > th | dose dependent | rect. | 21 | 12 |
| 630 | 71.25 | temperature | 10 | 4320 | > th | dose dependent | sin | 10 | NA |
| 631 | 71.25 | temperature | 30 | 4320 | < th | dose dependent | rect. | 3 | 1 |
| 640 | 71.26 | temperature | 10 | 4320 | > th | dose dependent | rect. | 3 | 1 |
| 711 | 71.37 | temperature | 20 | 43200 | < th | dose independent | rect. | 19 | 3 |
| 737 | 71.41 | temperature | 30 | 4320 | < th | dose dependent | sin | 10 | NA |
| 823 | 71.55 | temperature | 20 | 43200 | < th | dose independent | rect. | 20 | 1 |
| 877 | 71.61 | wind | 5 | 43200 | > th | dose dependent | rect. | 10 | 2 |
| 970 | 71.74 | radiation | 1 | 14400 | < th | dose independent | rect. | 24 | 7 |
| 972 | 71.74 | radiation | 1 | 14400 | < th | dose independent | rect. | 21 | 10 |