Os10g0204000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description :
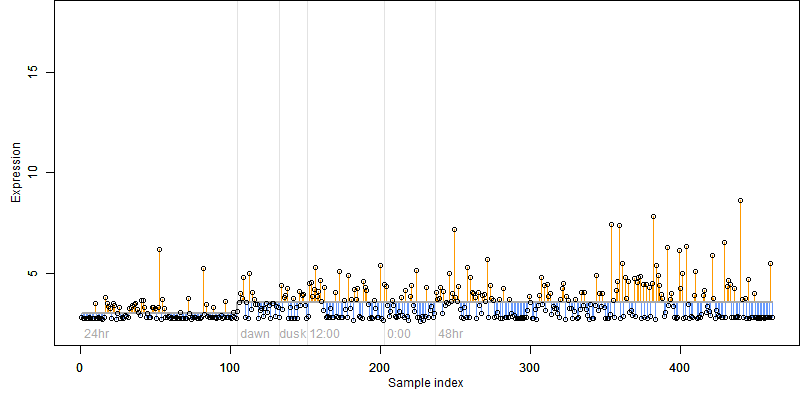

FiT-DB / Search/ Help/ Sample detail
|
Os10g0204000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description :
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
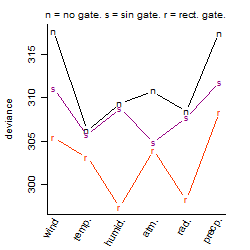
Dependence on each variable

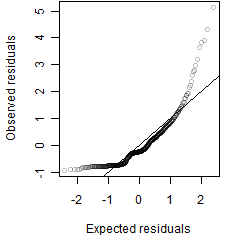
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 308.01 | 0.825 | 3.37 | 0.734 | 0.161 | 0.346 | -0.985 | 4.02 | -0.311 | 8.22 | 15 | 4421 | -- | -- |
| 317.35 | 0.83 | 3.5 | -0.225 | 0.201 | 0.456 | -0.856 | -- | -0.462 | 8.16 | 29.82 | 4108 | -- | -- |
| 310.64 | 0.821 | 3.36 | 0.739 | 0.177 | 0.332 | -- | 4 | -0.302 | 8.78 | 15.65 | 4824 | -- | -- |
| 319.44 | 0.832 | 3.51 | -0.286 | 0.177 | 0.472 | -- | -- | -0.508 | 8.99 | 27.08 | 4100 | -- | -- |
| 312.37 | 0.823 | 3.37 | 0.744 | -- | 0.304 | -- | 3.96 | -0.295 | -- | 15.88 | 4809 | -- | -- |
| 324.08 | 0.844 | 3.51 | -0.122 | 0.158 | -- | -0.904 | -- | -0.531 | 8.31 | -- | -- | -- | -- |
| 326.63 | 0.846 | 3.51 | -0.148 | 0.159 | -- | -- | -- | -0.531 | 8.52 | -- | -- | -- | -- |
| 321.28 | 0.835 | 3.51 | -0.281 | -- | 0.457 | -- | -- | -0.513 | -- | 27 | 4153 | -- | -- |
| 321.25 | 0.835 | 3.49 | -- | 0.174 | 0.407 | -- | -- | -0.436 | 9.42 | 29.7 | 4109 | -- | -- |
| 328.05 | 0.846 | 3.52 | -0.148 | -- | -- | -- | -- | -0.529 | -- | -- | -- | -- | -- |
| 327.21 | 0.846 | 3.51 | -- | 0.159 | -- | -- | -- | -0.514 | 8.59 | -- | -- | -- | -- |
| 322.99 | 0.837 | 3.5 | -- | -- | 0.393 | -- | -- | -0.436 | -- | 29.8 | 4139 | -- | -- |
| 328.62 | 0.846 | 3.51 | -- | -- | -- | -- | -- | -0.513 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance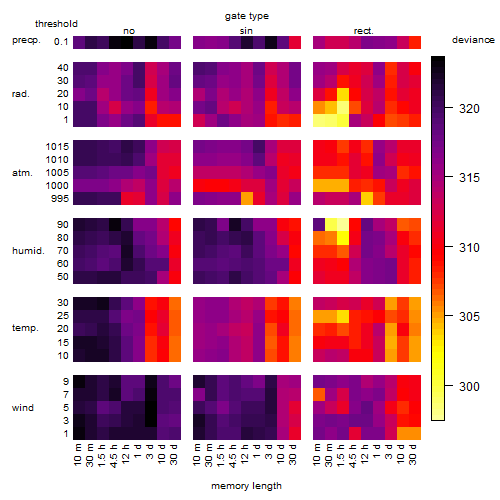
|
Histogram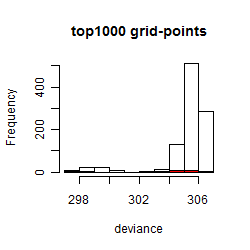 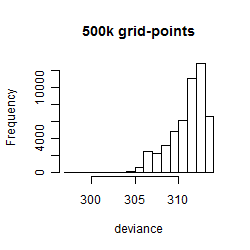
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 297.43 | humidity | 90 | 90 | > th | dose independent | rect. | 14 | 1 |
| 4 | 297.43 | humidity | 90 | 90 | > th | dose dependent | rect. | 14 | 1 |
| 7 | 298.32 | radiation | 1 | 10 | < th | dose independent | rect. | 15 | 1 |
| 11 | 298.46 | radiation | 1 | 10 | < th | dose independent | rect. | 11 | 5 |
| 23 | 298.61 | radiation | 10 | 90 | < th | dose dependent | rect. | 14 | 1 |
| 64 | 303.18 | temperature | 25 | 90 | > th | dose independent | rect. | 1 | 1 |
| 66 | 303.39 | radiation | 1 | 10 | < th | dose dependent | rect. | 15 | 1 |
| 77 | 303.99 | atmosphere | 995 | 720 | < th | dose dependent | rect. | 18 | 22 |
| 81 | 304.32 | atmosphere | 995 | 720 | < th | dose independent | rect. | 18 | 22 |
| 92 | 304.66 | atmosphere | 995 | 720 | < th | dose independent | rect. | 14 | 1 |
| 133 | 304.66 | atmosphere | 995 | 720 | < th | dose dependent | rect. | 14 | 1 |
| 174 | 304.67 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 1 | 1 |
| 181 | 304.67 | atmosphere | 1000 | 10 | < th | dose dependent | rect. | 1 | 1 |
| 192 | 304.75 | temperature | 25 | 90 | > th | dose dependent | rect. | 1 | 1 |
| 193 | 304.77 | temperature | 15 | 4320 | > th | dose dependent | rect. | 16 | 1 |
| 195 | 304.83 | atmosphere | 995 | 720 | > th | dose independent | sin | 7 | NA |
| 205 | 305.00 | temperature | 30 | 4320 | < th | dose dependent | rect. | 16 | 1 |
| 209 | 305.19 | temperature | 15 | 43200 | > th | dose dependent | rect. | 19 | 1 |
| 211 | 305.21 | temperature | 30 | 43200 | < th | dose dependent | rect. | 19 | 1 |
| 227 | 305.32 | temperature | 15 | 43200 | > th | dose dependent | rect. | 21 | 1 |
| 237 | 305.34 | temperature | 30 | 43200 | < th | dose dependent | rect. | 21 | 1 |
| 278 | 305.47 | wind | 1 | 43200 | < th | dose independent | rect. | 2 | 1 |
| 299 | 305.51 | wind | 1 | 14400 | > th | dose independent | rect. | 2 | 1 |
| 381 | 305.67 | atmosphere | 995 | 720 | < th | dose independent | sin | 7 | NA |
| 445 | 305.75 | temperature | 30 | 43200 | < th | dose dependent | sin | 16 | NA |
| 488 | 305.81 | temperature | 10 | 43200 | > th | dose dependent | sin | 15 | NA |
| 747 | 306.03 | wind | 1 | 43200 | < th | dose independent | rect. | 2 | 3 |
| 798 | 306.06 | temperature | 25 | 43200 | < th | dose independent | rect. | 12 | 9 |