Os09g0560200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to 26S protease regulatory subunit 6B (MIP224) (MB67 interacting protein) (TAT-binding protein-7) (TBP-7). Splice isoform 2.

FiT-DB / Search/ Help/ Sample detail
|
Os09g0560200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to 26S protease regulatory subunit 6B (MIP224) (MB67 interacting protein) (TAT-binding protein-7) (TBP-7). Splice isoform 2.
|
|
log2(Expression) ~ Norm(μ, σ2)
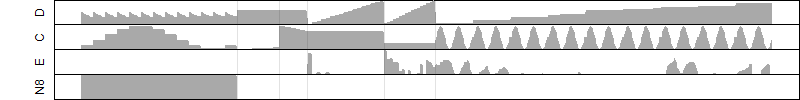
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable

Residual plot

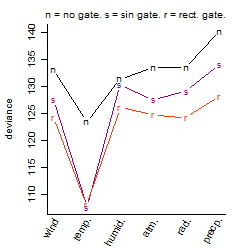
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 121.99 | 0.519 | 7.33 | 0.961 | 1.08 | 1.16 | -0.188 | -0.95 | -0.291 | 16.2 | 23.16 | 10 | -- | -- |
| 122.96 | 0.52 | 7.34 | 0.859 | 1.09 | 1.31 | 0.138 | -- | -0.307 | 16.1 | 23.16 | 10 | -- | -- |
| 121.83 | 0.518 | 7.33 | 0.952 | 1.08 | 1.18 | -- | -0.872 | -0.292 | 16.2 | 23.16 | 10 | -- | -- |
| 123 | 0.52 | 7.34 | 0.86 | 1.09 | 1.3 | -- | -- | -0.307 | 16.1 | 23.16 | 10 | -- | -- |
| 182.23 | 0.632 | 7.33 | 0.87 | -- | 0.68 | -- | -1.34 | -0.37 | -- | 23.16 | 10 | -- | -- |
| 144.26 | 0.563 | 7.39 | 0.593 | 0.98 | -- | -0.184 | -- | -0.482 | 16.8 | -- | -- | -- | -- |
| 144.33 | 0.563 | 7.39 | 0.593 | 0.978 | -- | -- | -- | -0.483 | 16.7 | -- | -- | -- | -- |
| 183.86 | 0.632 | 7.36 | 0.754 | -- | 1.29 | -- | -- | -0.424 | -- | 20.46 | 23 | -- | -- |
| 140.76 | 0.553 | 7.38 | -- | 1.05 | 1.02 | -- | -- | -0.451 | 16.3 | 22.5 | 10 | -- | -- |
| 195.02 | 0.653 | 7.38 | 0.556 | -- | -- | -- | -- | -0.508 | -- | -- | -- | -- | -- |
| 153.6 | 0.58 | 7.4 | -- | 0.967 | -- | -- | -- | -0.549 | 16.7 | -- | -- | -- | -- |
| 197.44 | 0.654 | 7.38 | -- | -- | 0.812 | -- | -- | -0.524 | -- | 20.78 | 18 | -- | -- |
| 203.17 | 0.665 | 7.39 | -- | -- | -- | -- | -- | -0.57 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 107.68 | temperature | 25 | 10 | < th | dose dependent | sin | 14 | NA |
| 3 | 108.16 | temperature | 25 | 90 | < th | dose dependent | rect. | 15 | 12 |
| 4 | 108.16 | temperature | 25 | 90 | < th | dose dependent | rect. | 13 | 14 |
| 5 | 108.26 | temperature | 25 | 10 | < th | dose dependent | rect. | 13 | 13 |
| 9 | 108.36 | temperature | 25 | 10 | < th | dose dependent | rect. | 16 | 10 |
| 94 | 111.74 | temperature | 25 | 10 | < th | dose dependent | rect. | 16 | 14 |
| 145 | 112.62 | temperature | 10 | 720 | > th | dose dependent | rect. | 16 | 7 |
| 361 | 115.75 | temperature | 25 | 720 | < th | dose dependent | rect. | 19 | 1 |
| 371 | 115.90 | temperature | 10 | 10 | > th | dose dependent | sin | 12 | NA |
| 422 | 116.54 | temperature | 25 | 270 | < th | dose dependent | rect. | 21 | 2 |
| 458 | 116.84 | temperature | 20 | 30 | < th | dose independent | sin | 14 | NA |
| 542 | 117.47 | temperature | 15 | 270 | > th | dose dependent | rect. | 16 | 12 |
| 558 | 117.54 | temperature | 25 | 270 | < th | dose independent | rect. | 24 | 11 |
| 615 | 118.02 | temperature | 20 | 270 | > th | dose independent | rect. | 15 | 13 |
| 628 | 118.09 | temperature | 20 | 90 | < th | dose independent | rect. | 16 | 11 |
| 651 | 118.26 | temperature | 20 | 90 | < th | dose independent | rect. | 12 | 15 |
| 664 | 118.34 | temperature | 20 | 90 | > th | dose independent | rect. | 17 | 14 |
| 672 | 118.42 | temperature | 20 | 90 | > th | dose independent | sin | 14 | NA |
| 743 | 118.84 | temperature | 30 | 720 | < th | dose dependent | rect. | 13 | 2 |
| 891 | 119.30 | temperature | 25 | 720 | < th | dose independent | rect. | 12 | 10 |
| 911 | 119.38 | temperature | 20 | 30 | < th | dose independent | rect. | 16 | 14 |
| 917 | 119.40 | temperature | 20 | 30 | < th | dose independent | rect. | 12 | 18 |
| 971 | 119.66 | temperature | 20 | 90 | < th | dose independent | rect. | 19 | 8 |